NMR-based Stable Isotope Resolved Metabolomics in systems biochemistry
- PMID: 28263717
- PMCID: PMC5545158
- DOI: 10.1016/j.abb.2017.02.009
NMR-based Stable Isotope Resolved Metabolomics in systems biochemistry
Abstract
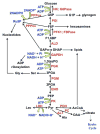
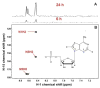
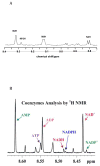
Metabolism is the basic activity of live cells, and monitoring the metabolic state provides a dynamic picture of the cells or tissues, and how they respond to external changes, for in disease or treatment with drugs. NMR is an extremely versatile analytical tool that can be applied to a wide range of biochemical problems. Despite its modest sensitivity its versatility make it an ideal tool for analyzing biochemical dynamics both in vitro and in vivo, especially when coupled with its isotope editing capabilities, from which isotope distributions can be readily determined. These are critical for any analyses of flux in live organisms. This review focuses on the utility of NMR spectroscopy in metabolomics, with an emphasis on NMR applications in stable isotope-enriched tracer research for elucidating biochemical pathways and networks with examples from nucleotide biochemistry. The knowledge gained from this area of research provides a ready link to genomic, epigenomic, transcriptomic, and proteomic information to achieve systems biochemical understanding of living cells and organisms.
Keywords: Isotope editing; Isotopomer distribution analysis; Stable Isotope Resolved Metabolomics (SIRM).
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
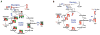
Similar articles
-
Applications of NMR spectroscopy to systems biochemistry.Prog Nucl Magn Reson Spectrosc. 2016 Feb;92-93:18-53. doi: 10.1016/j.pnmrs.2016.01.005. Epub 2016 Feb 6. Prog Nucl Magn Reson Spectrosc. 2016. PMID: 26952191 Free PMC article. Review.
-
NMR-Based Stable Isotope Tracing of Cancer Metabolism.Methods Mol Biol. 2025;2855:457-504. doi: 10.1007/978-1-0716-4116-3_26. Methods Mol Biol. 2025. PMID: 39354323
-
NMR-based isotope editing, chemoselection and isotopomer distribution analysis in stable isotope resolved metabolomics.Methods. 2022 Oct;206:8-17. doi: 10.1016/j.ymeth.2022.07.014. Epub 2022 Jul 28. Methods. 2022. PMID: 35908585 Free PMC article. Review.
-
Exploring cancer metabolism using stable isotope-resolved metabolomics (SIRM).J Biol Chem. 2017 Jul 14;292(28):11601-11609. doi: 10.1074/jbc.R117.776054. Epub 2017 Jun 7. J Biol Chem. 2017. PMID: 28592486 Free PMC article. Review.
-
NMR-based stable isotope resolved metabolomics in systems biochemistry.J Biomol NMR. 2011 Apr;49(3-4):267-80. doi: 10.1007/s10858-011-9484-6. Epub 2011 Feb 26. J Biomol NMR. 2011. PMID: 21350847 Free PMC article.
Cited by
-
Targeting Metabolic Cross Talk Between Cancer Cells and Cancer-Associated Fibroblasts.Adv Exp Med Biol. 2021;1311:205-214. doi: 10.1007/978-3-030-65768-0_15. Adv Exp Med Biol. 2021. PMID: 34014545 Free PMC article.
-
Metabolomic Approaches to Study Chemical Exposure-Related Metabolism Alterations in Mammalian Cell Cultures.Int J Mol Sci. 2020 Sep 18;21(18):6843. doi: 10.3390/ijms21186843. Int J Mol Sci. 2020. PMID: 32961865 Free PMC article. Review.
-
Challenges of Spatially Resolved Metabolism in Cancer Research.Metabolites. 2024 Jul 11;14(7):383. doi: 10.3390/metabo14070383. Metabolites. 2024. PMID: 39057706 Free PMC article. Review.
-
NMR Analysis of Carboxylate Isotopomers of 13C-Metabolites by Chemoselective Derivatization with 15N-Cholamine.Anal Chem. 2021 May 4;93(17):6629-6637. doi: 10.1021/acs.analchem.0c04220. Epub 2021 Apr 21. Anal Chem. 2021. PMID: 33880916 Free PMC article.
-
Applications of Chromatography-Ultra High-Resolution MS for Stable Isotope-Resolved Metabolomics (SIRM) Reconstruction of Metabolic Networks.Trends Analyt Chem. 2020 Feb;123:115676. doi: 10.1016/j.trac.2019.115676. Epub 2019 Oct 1. Trends Analyt Chem. 2020. PMID: 32483395 Free PMC article.
References
-
- Schanda P, Meier BH, Ernst M. Quantitative Analysis of Protein Backbone Dynamics in Microcrystalline Ubiquitin by Solid-State NMR Spectroscopy. J Am Chem Soc. 2010;132:15957–15967. - PubMed
-
- Levitt MH. Spin Dynamics: Basics of Nuclear Magnetic Resonance. Chichester, United Kingdom: Wiley;
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

