Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells
- PMID: 28264033
- PMCID: PMC5354447
- DOI: 10.1371/journal.ppat.1006265
Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells
Abstract
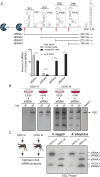
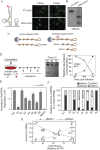
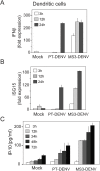
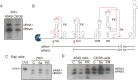
The Flavivirus genus includes a large number of medically relevant pathogens that cycle between humans and arthropods. This host alternation imposes a selective pressure on the viral population. Here, we found that dengue virus, the most important viral human pathogen transmitted by insects, evolved a mechanism to differentially regulate the production of viral non-coding RNAs in mosquitos and humans, with a significant impact on viral fitness in each host. Flavivirus infections accumulate non-coding RNAs derived from the viral 3'UTRs (known as sfRNAs), relevant in viral pathogenesis and immune evasion. We found that dengue virus host adaptation leads to the accumulation of different species of sfRNAs in vertebrate and invertebrate cells. This process does not depend on differences in the host machinery; but it was found to be dependent on the selection of specific mutations in the viral 3'UTR. Dissecting the viral population and studying phenotypes of cloned variants, the molecular determinants for the switch in the sfRNA pattern during host change were mapped to a single RNA structure. Point mutations selected in mosquito cells were sufficient to change the pattern of sfRNAs, induce higher type I interferon responses and reduce viral fitness in human cells, explaining the rapid clearance of certain viral variants after host change. In addition, using epidemic and pre-epidemic Zika viruses, similar patterns of sfRNAs were observed in mosquito and human infected cells, but they were different from those observed during dengue virus infections, indicating that distinct selective pressures act on the 3'UTR of these closely related viruses. In summary, we present a novel mechanism by which dengue virus evolved an RNA structure that is under strong selective pressure in the two hosts, as regulator of non-coding RNA accumulation and viral fitness. This work provides new ideas about the impact of host adaptation on the variability and evolution of flavivirus 3'UTRs with possible implications in virulence and viral transmission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rico-Hesse R (1990) Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174: 479–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

