Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA
- PMID: 28264529
- PMCID: PMC5372735
- DOI: 10.3390/biom7010023
Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA
Abstract
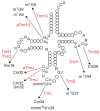
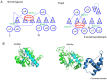
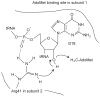
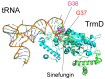
The existence of SpoU-TrmD (SPOUT) RNA methyltransferase superfamily was first predicted by bioinformatics. SpoU is the previous name of TrmH, which catalyzes the 2'-Omethylation of ribose of G18 in tRNA; TrmD catalyzes the formation of N1-methylguanosine at position 37 in tRNA. Although SpoU (TrmH) and TrmD were originally considered to be unrelated, the bioinformatics study suggested that they might share a common evolution origin and form a single superfamily. The common feature of SPOUT RNA methyltransferases is the formation of a deep trefoil knot in the catalytic domain. In the past decade, the SPOUT RNA methyltransferase superfamily has grown; furthermore, knowledge concerning the functions of their modified nucleosides in tRNA has also increased. Some enzymes are potential targets in the design of antibacterial drugs. In humans, defects in some genes may be related to carcinogenesis. In this review, recent findings on the tRNA methyltransferases with a SPOUT fold and their methylated nucleosides in tRNA, including classification of tRNA methyltransferases with a SPOUT fold; knot structures, domain arrangements, subunit structures and reaction mechanisms; tRNA recognition mechanisms, and functions of modified nucleosides synthesized by this superfamily, are summarized. Lastly, the future perspective for studies on tRNA modification enzymes are considered.
Keywords: knot; RNA modification; SpoU‐TrmD; methyltransferase; tRNA.
Conflict of interest statement
The author declares no conflict of interest.
Figures



Similar articles
-
Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):E4197-205. doi: 10.1073/pnas.1422981112. Epub 2015 Jul 16. Proc Natl Acad Sci U S A. 2015. PMID: 26183229 Free PMC article.
-
Tied up in knots: Untangling substrate recognition by the SPOUT methyltransferases.J Biol Chem. 2022 Oct;298(10):102393. doi: 10.1016/j.jbc.2022.102393. Epub 2022 Aug 18. J Biol Chem. 2022. PMID: 35988649 Free PMC article. Review.
-
A Family Divided: Distinct Structural and Mechanistic Features of the SpoU-TrmD (SPOUT) Methyltransferase Superfamily.Biochemistry. 2019 Feb 5;58(5):336-345. doi: 10.1021/acs.biochem.8b01047. Epub 2018 Dec 3. Biochemistry. 2019. PMID: 30457841 Free PMC article.
-
Mechanistic features of the atypical tRNA m1G9 SPOUT methyltransferase, Trm10.Nucleic Acids Res. 2017 Sep 6;45(15):9019-9029. doi: 10.1093/nar/gkx620. Nucleic Acids Res. 2017. PMID: 28911116 Free PMC article.
-
Trm5 and TrmD: Two Enzymes from Distinct Origins Catalyze the Identical tRNA Modification, m¹G37.Biomolecules. 2017 Mar 21;7(1):32. doi: 10.3390/biom7010032. Biomolecules. 2017. PMID: 28335556 Free PMC article. Review.
Cited by
-
Arabidopsis TRM5 encodes a nuclear-localised bifunctional tRNA guanine and inosine-N1-methyltransferase that is important for growth.PLoS One. 2019 Nov 22;14(11):e0225064. doi: 10.1371/journal.pone.0225064. eCollection 2019. PLoS One. 2019. PMID: 31756231 Free PMC article.
-
Genetic dissection of marker trait associations for grain micro-nutrients and thousand grain weight under heat and drought stress conditions in wheat.Front Plant Sci. 2023 Jan 16;13:1082513. doi: 10.3389/fpls.2022.1082513. eCollection 2022. Front Plant Sci. 2023. PMID: 36726675 Free PMC article.
-
Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA.Genes (Basel). 2023 Jan 31;14(2):382. doi: 10.3390/genes14020382. Genes (Basel). 2023. PMID: 36833309 Free PMC article. Review.
-
Potential therapeutic targets from Mycobacterium abscessus (Mab): recently reported efforts towards the discovery of novel antibacterial agents to treat Mab infections.RSC Med Chem. 2022 Mar 10;13(4):392-404. doi: 10.1039/d1md00359c. eCollection 2022 Apr 20. RSC Med Chem. 2022. PMID: 35647542 Free PMC article. Review.
-
Codon-Specific Translation by m1G37 Methylation of tRNA.Front Genet. 2019 Jan 10;9:713. doi: 10.3389/fgene.2018.00713. eCollection 2018. Front Genet. 2019. PMID: 30687389 Free PMC article.
References
-
- Anantharaman V., Koonin E.V., Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

