A Family of Single Copy repABC-Type Shuttle Vectors Stably Maintained in the Alpha-Proteobacterium Sinorhizobium meliloti
- PMID: 28264559
- PMCID: PMC7610768
- DOI: 10.1021/acssynbio.6b00320
A Family of Single Copy repABC-Type Shuttle Vectors Stably Maintained in the Alpha-Proteobacterium Sinorhizobium meliloti
Abstract
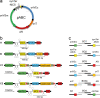
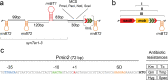
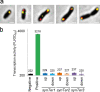
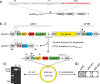
A considerable share of bacterial species maintains segmented genomes. Plant symbiotic α-proteobacterial rhizobia contain up to six repABC-type replicons in addition to the primary chromosome. These low or unit-copy replicons, classified as secondary chromosomes, chromids, or megaplasmids, are exclusively found in α-proteobacteria. Replication and faithful partitioning of these replicons to the daughter cells is mediated by the repABC region. The importance of α-rhizobial symbiotic nitrogen fixation for sustainable agriculture and Agrobacterium-mediated plant transformation as a tool in plant sciences has increasingly moved biological engineering of these organisms into focus. Plasmids are ideal DNA-carrying vectors for these engineering efforts. On the basis of repABC regions collected from α-rhizobial secondary replicons, and origins of replication derived from traditional cloning vectors, we devised the versatile family of pABC shuttle vectors propagating in Sinorhizobium meliloti, related members of the Rhizobiales, and Escherichia coli. A modular plasmid library providing the elemental parts for pABC vector assembly was founded. The standardized design of these vectors involves five basic modules: (1) repABC cassette, (2) plasmid-derived origin of replication, (3) RK2/RP4 mobilization site (optional), (4) antibiotic resistance gene, and (5) multiple cloning site flanked by transcription terminators. In S. meliloti, pABC vectors showed high propagation stability and unit-copy number. We demonstrated stable coexistence of three pABC vectors in addition to the two indigenous megaplasmids in S. meliloti, suggesting combinability of multiple compatible pABC plasmids. We further devised an in vivo cloning strategy involving Cre/lox-mediated translocation of large DNA fragments to an autonomously replicating repABC-based vector, followed by conjugation-mediated transfer either to compatible rhizobia or E. coli.
Keywords: Cre/loxP; Rhizobiales; Sinorhizobium; in vivo cloning; plasmid cloning vehicle; repABC.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids.Mol Microbiol. 2005 Jan;55(2):611-23. doi: 10.1111/j.1365-2958.2004.04412.x. Mol Microbiol. 2005. PMID: 15659174
-
Differential Localization and Functional Specialization of parS Centromere-Like Sites in repABC Replicons of Alphaproteobacteria.Appl Environ Microbiol. 2022 Apr 26;88(8):e0020722. doi: 10.1128/aem.00207-22. Epub 2022 Apr 7. Appl Environ Microbiol. 2022. PMID: 35389251 Free PMC article.
-
Identification of a megaplasmid centromere reveals genetic structural diversity within the repABC family of basic replicons.Mol Microbiol. 2006 Mar;59(5):1559-75. doi: 10.1111/j.1365-2958.2006.05040.x. Mol Microbiol. 2006. PMID: 16468995
-
The ABCs of plasmid replication and segregation.Nat Rev Microbiol. 2012 Nov;10(11):755-65. doi: 10.1038/nrmicro2882. Nat Rev Microbiol. 2012. PMID: 23070556 Review.
-
The Plasmid Mobilome of the Model Plant-Symbiont Sinorhizobium meliloti: Coming up with New Questions and Answers.Microbiol Spectr. 2014 Oct;2(5). doi: 10.1128/microbiolspec.PLAS-0005-2013. Microbiol Spectr. 2014. PMID: 26104371 Review.
Cited by
-
Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae.ISME J. 2018 Aug;12(8):1994-2010. doi: 10.1038/s41396-018-0150-9. Epub 2018 May 24. ISME J. 2018. PMID: 29795276 Free PMC article.
-
Genome-wide comparative analysis of clinical and environmental strains of the opportunistic pathogen Paracoccus yeei (Alphaproteobacteria).Front Microbiol. 2024 Nov 6;15:1483110. doi: 10.3389/fmicb.2024.1483110. eCollection 2024. Front Microbiol. 2024. PMID: 39568992 Free PMC article.
-
An Integrated Affinity Chromatography-Based Approach to Unravel the sRNA Interactome in Nitrogen-Fixing Rhizobia.Methods Mol Biol. 2024;2741:363-380. doi: 10.1007/978-1-0716-3565-0_19. Methods Mol Biol. 2024. PMID: 38217663
-
Seven-transmembrane receptor protein RgsP and cell wall-binding protein RgsM promote unipolar growth in Rhizobiales.PLoS Genet. 2018 Aug 13;14(8):e1007594. doi: 10.1371/journal.pgen.1007594. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30102748 Free PMC article.
-
Design and Control of Extrachromosomal Elements in Methylorubrum extorquens AM1.ACS Synth Biol. 2019 Nov 15;8(11):2451-2456. doi: 10.1021/acssynbio.9b00220. Epub 2019 Oct 21. ACS Synth Biol. 2019. PMID: 31584803 Free PMC article.
References
-
- Gerdes K.; Howard M.; Szardenings F. (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

