Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding
- PMID: 28264564
- PMCID: PMC5724935
- DOI: 10.1021/acssynbio.6b00309
Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding
Abstract
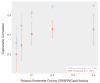
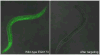
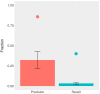
Lineage tracing by the determination and mapping of progeny arising from single cells is an important approach enabling the elucidation of mechanisms underlying diverse biological processes ranging from development to disease. We developed a dynamic sequence-based barcode system for synthetic lineage tracing and have demonstrated its performance in C. elegans, a model organism whose lineage tree is well established. The strategy we use creates lineage trees based upon the introduction of synthetically controlled mutations into cells and the propagation of these mutations to daughter cells at each cell division. We analyzed this experimental proof of concept along with a corresponding simulation and analytical model to gain a deeper understanding of the coding capacity of the system. Our results provide specific bounds on the fidelity of lineage tracing using such approaches.
Keywords: CRISPR/Cas9; DNA barcoding; lineage tracing.
Figures



References
-
- Kretzschmar K, Watt FM. Lineage Tracing. Cell. 2012;148:33–45. - PubMed
-
- Wilson HV. Embryology of the Sea Bass. Government Printing Office; Washington, D.C: 1891.
-
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. - PubMed
-
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. - PubMed
-
- Vogt W. Teil Gastrulation und Mesodermbildung bei Urodelen und Anuren. Gestaltungsanalyse am Amphibienkeim mit örtlicher Vitalfärbung. Dev. Genes Evol. 1929;120:384–706. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

