Insight into SNPs and epitopes of E protein of newly emerged genotype-I isolates of JEV from Midnapur, West Bengal, India
- PMID: 28264652
- PMCID: PMC5339996
- DOI: 10.1186/s12865-017-0197-9
Insight into SNPs and epitopes of E protein of newly emerged genotype-I isolates of JEV from Midnapur, West Bengal, India
Abstract
Background: Japanese encephalitis virus (JEV) is a mosquito-borne flavivirus that causes Japanese Encephalitis (JE) and Acute Encephalitis Syndrome (AES) in humans. Genotype-I (as co-circulating cases with Genotype-III) was isolated in 2010 (JEV28, JEV21) and then in 2011 (JEV45) from Midnapur district, West Bengal (WB) for the first time from clinical patients who were previously been vaccinated with live attenuated SA14-14-2 strain. We apply bioinformatics and immunoinformatics on sequence and structure of E protein for analysis of crucial substitutions that might cause the genotypic transition, affecting protein-function and altering specificity of epitopes.
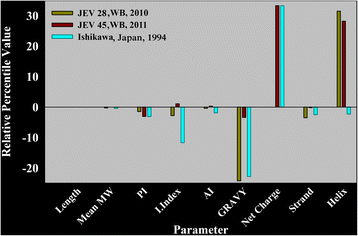
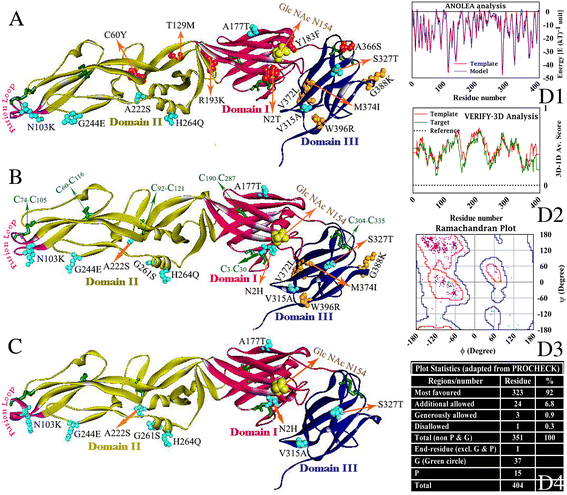
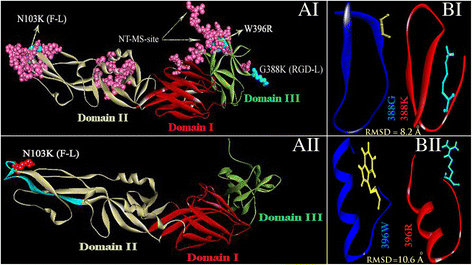
Results: Although frequency of substitutions in E glycoprotein of JEV28, JEV21 and JEV45 isolates vary, its homologous patterns remain exactly similar as earlier Japan isolate (Ishikawa). Sequence and 3D model-structure based analyses of E protein show that only four of all substitutions are critical for genotype-I specific effect of which N103K is common among all isolates indicating its role in the transition of genotype-III to genotype-I. Predicted B-cell and T-cell epitopes are seen to harbor these critical substitutions that affect overall conformational stability of the protein. These epitopes were subjected to conservation analyses using a large set of the protein from Asian continent.
Conclusions: The study identifies crucial substitutions that contribute to the emergence of genotype-I. Predicted epitopes harboring these substitutions may alter specificity which might be the reason of reported failure of vaccine. Conservation analysis of these epitopes would be useful for design of genotype-I specific vaccine.
Keywords: B-cell & T-cell epitopes; Docking; Genotype I; Genotype III; Homology model; Japanese encephalitis virus; Midnapur; PEP-FOLD; SNP Energetics.
Figures


References
-
- Saxena SK, Mishra N, Saxena R, Singh M, Mathur A. Trend of Japanese encephalitis in North India: evidence from thirty-eight acute encephalitis cases and appraisal of niceties. J Infect Dev Ctries. 2009;30:517–530. - PubMed
-
- Sarkar A, Banerjee S, Mukhopadhyay SK, Chatterjee S. Etiological spectrum of co-circulating Japanese encephalitis virus genotype I and III in clinically diagnosed AES cases from West Bengal, India: an indication of public health threat in near future. Int J Institutional Pharmacy Life Sci. 2015;5:154-180.
-
- Sarkar A, Taraphdar D, Mukhopadhyay SK, Chakrabarti S, Chatterjee S. Molecular evidence for the occurrence of Japanese encephalitis virus genotype I and III infection associated with acute encephalitis in patients of West Bengal, India, 2010. Virol J. 2012;9:271. doi: 10.1186/1743-422X-9-271. - DOI - PMC - PubMed
-
- Sarkar A, Banik A, Pathak BK, Mukhopadhyay SK, Chatterjee S. Envelope protein gene based molecular characterization of Japanese encephalitis virus clinical isolates from West Bengal, India: a comparative approach with respect to SA14-14-2 live attenuated vaccine strain. BMC Infect Dis. 2013;13:368. doi: 10.1186/1471-2334-13-368. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

