Promotion and Rescue of Intracellular Brucella neotomae Replication during Coinfection with Legionella pneumophila
- PMID: 28264909
- PMCID: PMC5400850
- DOI: 10.1128/IAI.00991-16
Promotion and Rescue of Intracellular Brucella neotomae Replication during Coinfection with Legionella pneumophila
Abstract
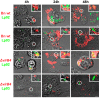
We established a new Brucella neotomaein vitro model system for study of type IV secretion system-dependent (T4SS) pathogenesis in the Brucella genus. Importantly, B. neotomae is a rodent pathogen, and unlike B. abortus, B. melitensis, and B. suis, B. neotomae has not been observed to infect humans. It therefore can be handled more facilely using biosafety level 2 practices. More particularly, using a series of novel fluorescent protein and lux operon reporter systems to differentially label pathogens and track intracellular replication, we confirmed T4SS-dependent intracellular growth of B. neotomae in macrophage cell lines. Furthermore, B. neotomae exhibited early endosomal (LAMP-1) and late endoplasmic reticulum (calreticulin)-associated phagosome maturation. These findings recapitulate prior observations for human-pathogenic Brucella spp. In addition, during coinfection experiments with Legionella pneumophila, we found that defective intracellular replication of a B. neotomae T4SS virB4 mutant was rescued and baseline levels of intracellular replication of wild-type B. neotomae were significantly stimulated by coinfection with wild-type but not T4SS mutant L. pneumophila Using confocal microscopy, it was determined that intracellular colocalization of B. neotomae and L. pneumophila was required for rescue and that colocalization came at a cost to L. pneumophila fitness. These findings were not completely expected based on known temporal and qualitative differences in the intracellular life cycles of these two pathogens. Taken together, we have developed a new system for studying in vitroBrucella pathogenesis and found a remarkable T4SS-dependent interplay between Brucella and Legionella during macrophage coinfection.
Keywords: Brucella; Legionella pneumophila; coinfection; fluorescent image analysis; intracellular pathogens; pathogenesis; phagosomes; reporter genes; type IV secretion system.
Copyright © 2017 American Society for Microbiology.
Figures
Similar articles
-
A Chemical Genetics Screen Reveals Influence of p38 Mitogen-Activated Protein Kinase and Autophagy on Phagosome Development and Intracellular Replication of Brucella neotomae in Macrophages.Infect Immun. 2019 Jul 23;87(8):e00044-19. doi: 10.1128/IAI.00044-19. Print 2019 Aug. Infect Immun. 2019. PMID: 31160361 Free PMC article.
-
Characterization of Legionella pneumophila pmiA, a gene essential for infectivity of protozoa and macrophages.Infect Immun. 2005 Oct;73(10):6272-82. doi: 10.1128/IAI.73.10.6272-6282.2005. Infect Immun. 2005. PMID: 16177298 Free PMC article.
-
Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake.Mol Microbiol. 1998 May;28(3):663-74. doi: 10.1046/j.1365-2958.1998.00841.x. Mol Microbiol. 1998. PMID: 9632267
-
Fatal attraction of mammalian cells to Legionella pneumophila.Mol Microbiol. 1998 Nov;30(4):689-95. doi: 10.1046/j.1365-2958.1998.01092.x. Mol Microbiol. 1998. PMID: 10094618 Review.
-
Molecular and cell biology of Legionella pneumophila.Int J Med Microbiol. 2004 Apr;293(7-8):519-27. doi: 10.1078/1438-4221-00286. Int J Med Microbiol. 2004. PMID: 15149027 Review.
Cited by
-
From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella.Front Cell Infect Microbiol. 2017 Nov 30;7:477. doi: 10.3389/fcimb.2017.00477. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29250488 Free PMC article. Review.
-
Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.PLoS Biol. 2023 May 16;21(5):e3002091. doi: 10.1371/journal.pbio.3002091. eCollection 2023 May. PLoS Biol. 2023. PMID: 37192172 Free PMC article.
-
Host-bacteria interactions: ecological and evolutionary insights from ancient, professional endosymbionts.FEMS Microbiol Rev. 2024 Jun 20;48(4):fuae021. doi: 10.1093/femsre/fuae021. FEMS Microbiol Rev. 2024. PMID: 39081075 Free PMC article. Review.
-
Uncovering the Hidden Credentials of Brucella Virulence.Microbiol Mol Biol Rev. 2021 Feb 10;85(1):e00021-19. doi: 10.1128/MMBR.00021-19. Print 2021 Feb 17. Microbiol Mol Biol Rev. 2021. PMID: 33568459 Free PMC article. Review.
-
Brucella neotomae Recapitulates Attributes of Zoonotic Human Disease in a Murine Infection Model.Infect Immun. 2018 Dec 19;87(1):e00255-18. doi: 10.1128/IAI.00255-18. Print 2019 Jan. Infect Immun. 2018. PMID: 30373892 Free PMC article.
References
-
- Dalrymple-Champneys W. 1960. Brucella infection and undulant fever in man. Oxford University Press, London, United Kingdom.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

