Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations
- PMID: 28264936
- PMCID: PMC5376749
- DOI: 10.15252/emmm.201606660
Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations
Abstract
Heterozygous inactivating mutations in ribosomal protein genes (RPGs) are associated with hematopoietic and developmental abnormalities, activation of p53, and altered risk of cancer in humans and model organisms. Here we performed a large-scale analysis of cancer genome data to examine the frequency and selective pressure of RPG lesions across human cancers. We found that hemizygous RPG deletions are common, occurring in about 43% of 10,744 cancer specimens and cell lines. Consistent with p53-dependent negative selection, such lesions are underrepresented in TP53-intact tumors (P ≪ 10-10), and shRNA-mediated knockdown of RPGs activated p53 in TP53-wild-type cells. In contrast, we did not see negative selection of RPG deletions in TP53-mutant tumors. RPGs are conserved with respect to homozygous deletions, and shRNA screening data from 174 cell lines demonstrate that further suppression of hemizygously deleted RPGs inhibits cell growth. Our results establish RPG haploinsufficiency as a strikingly common vulnerability of human cancers that associates with TP53 mutations and could be targetable therapeutically.
Keywords: cancer; ribosomal gene haploinsufficiency; ribosome function.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
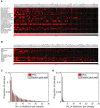
Deletion frequencies for different RPGs estimated from DNA copy number profiles of 7,275 primary cancer specimens belonging to 24 different tumor types (red).
Analysis of DNA copy number profiles of 2,476 primary cancer specimens belonging to 13 tumor types (including four not represented in the TCGA) identified the same RPGs as frequently deleted. Similar results were also calculated with DNA copy number profiles of 1,043 cancer cell lines (Fig EV1). The results shown were obtained using log2 ratio −0.5 as threshold. Similar results were obtained with other reasonable thresholds.
Histogram of number of deletions per tumor for RPGs and for 1,000 equally sized random gene sets in the TCGA samples. Tumors with multiple RPG deletions were common. Consistent with negative selection, we also detected fewer deletions in RPGs than in 1,000 random gene sets of the same size. This plot was obtained with a log2 threshold of −0.5, detecting both hemizygous and homozygous deletions.
Corresponding plot obtained with the more conservative log2 threshold −2.0, primarily detecting homozygous deletions.
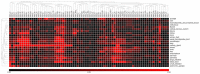
- A
RPG deletions are underrepresented in TP53‐intact tumors, but not in TP53‐mutant tumors. Plots show cumulative copy number distributions for RPGs (blue) versus all genes (dashed black) for 4,675 TCGA samples having both copy number microarray and whole‐exome sequence data. However, cases without TP53 mutation/deletion show fewer copy numbers consistent with heterozygous loss of RPGs compared to other genes (brackets), whereas cases harboring TP53 mutations/deletions show no corresponding difference. These plots also show that, regardless of TP53 mutation status, homozygous loss of RPGs is extremely rare.
- B
Number of hemizygous RPG deletions in TP53‐mutant and ‐wild‐type tumors. Boxes indicate medians and the first and third quartiles. Whiskers indicate first and third quartiles ±1.5 times the interquartile range. Notches indicate confidence intervals around the median. P‐values indicate significance by Wilcoxon rank‐sum test.
- C, D
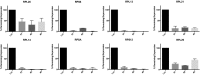
To obtain further support for p53‐negative selection, we used shRNA‐mediated knockdown of RPGs in A549 cells, which are TP53‐wild type. Panels show p53 protein levels as analyzed by Western blot (β‐actin loading control), and P21 transcript levels as analyzed by qPCR (ACTB normalization control) after 4–6 days of expression of two shRNAs per tested RPG. Error bars indicate standard deviation between triplicates. Throughout, we observed increased p53 levels and P21 expression. These data support that RPG deletions lead to p53 activation and negative selection in cancer cells.
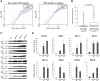
- A–C
Among tumors without detectable TP53 deletion/mutation, the RPG deletions that could be detected were enriched in tumors harboring alternative p53‐inactivating lesions, including MDM2 amplifications, CDKN2A deletions, and PTEN deletions. Results obtained using log2 ratio thresholds (A) −0.3; (B) −0.5; (C) −0.7. To avoid biases introduced by co‐deletion, RPGs on chromosome 12, 9, or 10 were excluded from the MDM2, CDKN2A, and PTEN analyses, respectively. Boxes indicate medians and the first and third quartiles. Whiskers indicate first and third quartiles ±1.5 times the interquartile range. Notches indicate confidence intervals around the median. P‐values indicate significance by Wilcoxon rank‐sum test.

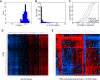
Correlations between RPG copy number and expression across 4,919 TCGA samples.
Numbers of RPG deletions per sample. Multiple RPG deletions are common.
Cumulative distributions of Pearson correlation coefficient for all pairs of RPGs across the TCGA RNA‐sequencing data. Tumors with more RPG copy number aberrations show less coordinated RPG expression.
Expression of RPGs in normal tissues (SymAtlas; NCBI Gene Expression Omnibus GSE96).
Expression of RPG in colorectal adenocarcinomas harboring 10 or more RPG copy number aberrations (TCGA samples).
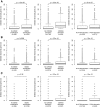
- A, B
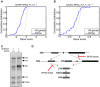
To investigate whether RPG haploinsufficiency renders cancer cells susceptible to further suppression of ribosome function, we analyzed genomewide pooled shRNA screening data from the (A) shARP and (B) Achilles studies (data for 72 and 102 genomically annotated cancer cell lines, respectively). For each targeted gene, the effect of knockdown on proliferation in cell lines carrying one gene copy, relative to those carrying at least two gene copies, was scored. We observed strongly significant enrichments of negative gene scores for RPGs (solid blue; P = 1.0 × 10−8 and P = 1.7 × 10−4, respectively) compared to other genes (gray), demonstrating that further suppression of hemizygously deleted RPGs inhibits cell growth.
- C, D
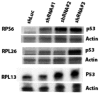
(C) To explore whether RPG haploinsufficiency causes rRNA maturation defects in cancer cells, we focused on pediatric ALL with 9p deletions targeting CDKN2A. From two previously banked samples (one with homozygous CDKN2A deletion but intact RPS6 and one with homozygous deletion of CDKN2A and hemizygous deletion of RPS6), we obtained viable cells, which were xenografted into NOD scid gamma immunodeficient mice and expanded in vivo. RNA from sorted tumor cells was assessed by Northern blot to analyze pre‐ribosomal RNA processing. The RPS6‐haploinsufficient case exhibited rRNA maturation defects similar to those seen previously in shRNA‐targeted RPS6‐deficient cells, namely (D) a reduction in 41S species (18S‐E, 21S and 41S) and an accumulation of 30S species (30S and 45S). These observations provide the first evidence indicating that the rRNA maturation defect that is observed in ribosomopathies may also be present in RPG‐haploinsufficient cancer cells.
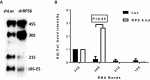
Representative Northern blot showing rRNA patterns in MOLM13 leukemia cells after knockdown with shRNA against RPS6 (shRPS6) or luciferase control (shLuc).
Quantification of rRNA specifies from three Northern blot gels (error bars show standard deviation; Student's t‐test P‐value). As shown, knockdown of RPS6 yielded increased 30S rRNA levels and decreased 21S and 18S rRNA levels, which is congruent with the results obtained for primary ALL cells shown in Fig 3.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

