KCNQ1 p.L353L affects splicing and modifies the phenotype in a founder population with long QT syndrome type 1
- PMID: 28264985
- PMCID: PMC5502312
- DOI: 10.1136/jmedgenet-2016-104153
KCNQ1 p.L353L affects splicing and modifies the phenotype in a founder population with long QT syndrome type 1
Abstract
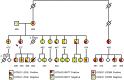
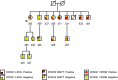
Background: Variable expressivity and incomplete penetrance between individuals with identical long QT syndrome (LQTS) causative mutations largely remain unexplained. Founder populations provide a unique opportunity to explore modifying genetic effects. We examined the role of a novel synonymous KCNQ1 p.L353L variant on the splicing of exon 8 and on heart rate corrected QT interval (QTc) in a population known to have a pathogenic LQTS type 1 (LQTS1) causative mutation, p.V205M, in KCNQ1-encoded Kv7.1.
Methods: 419 adults were genotyped for p.V205M, p.L353L and a previously described QTc modifier (KCNH2-p.K897T). Adjusted linear regression determined the effect of each variant on QTc, alone and in combination. In addition, peripheral blood RNA was extracted from three controls and three p.L353L-positive individuals. The mutant transcript levels were assessed via qPCR and normalised to overall KCNQ1 transcript levels to assess the effect on splicing.
Results: For women and men, respectively, p.L353L alone conferred a 10.0 (p=0.064) ms and 14.0 (p=0.014) ms increase in QTc and in men only a significant interaction effect in combination with the p.V205M (34.6 ms, p=0.003) resulting in a QTc of ∼500 ms. The mechanism of p.L353L's effect was attributed to approximately threefold increase in exon 8 exclusion resulting in ∼25% mutant transcripts of the total KCNQ1 transcript levels.
Conclusions: Our results provide the first evidence that synonymous variants outside the canonical splice sites in KCNQ1 can alter splicing and clinically impact phenotype. Through this mechanism, we identified that p.L353L can precipitate QT prolongation by itself and produce a clinically relevant interactive effect in conjunction with other LQTS variants.
Keywords: KCNQ1; First Nations; exon skipping; long QT syndrome; modifier.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/.
Conflict of interest statement
Competing interests: MJA is a consultant for Boston Scientific, Gilead Sciences, Medtronic and St. Jude Medical. MJA and Mayo Clinic receive royalties from Transgenomic for their FAMILION-LQTS and FAMILION-CPVT genetic tests. However, none of these commercial entities supported this research work.
Figures


References
-
- Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet 1996;12:17–23. 10.1038/ng0196-17 - DOI - PubMed
-
- Moss AJ, Shimizu W, Wilde AAM, Towbin JA, Zareba W, Robinson JL, Qi M, Vincent GM, Ackerman MJ, Kaufman ES, Hofman N, Seth R, Kamakura S, Miyamoto Y, Goldenberg I, Andrews ML, McNitt S. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation 2007;115:2481–9. 10.1161/CIRCULATIONAHA.106.665406 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
