Characterization of the catalytic properties of the membrane-anchored metalloproteinase ADAM9 in cell-based assays
- PMID: 28264989
- PMCID: PMC8606101
- DOI: 10.1042/BCJ20170075
Characterization of the catalytic properties of the membrane-anchored metalloproteinase ADAM9 in cell-based assays
Abstract
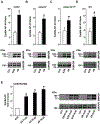
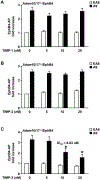
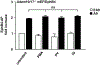
ADAM9 (A Disintegrin And Metalloprotease 9) is a membrane-anchored metalloproteinase that has been implicated in pathological retinal neovascularization and in tumor progression. ADAM9 has constitutive catalytic activity in both biochemical and cell-based assays and can cleave several membrane proteins, including epidermal growth factor and Ephrin receptor B4; yet little is currently known about the catalytic properties of ADAM9 and its post-translational regulation and inhibitor profile in cell-based assays. To address this question, we monitored processing of the membrane-anchored Ephrin receptor B4 (EphB4) by co-expressing ADAM9, with the catalytically inactive ADAM9 E > A mutant serving as a negative control. We found that ADAM9-dependent shedding of EphB4 was not stimulated by three commonly employed activators of ADAM-dependent ectodomain shedding: phorbol esters, pervanadate or calcium ionophores. With respect to the inhibitor profile, we found that ADAM9 was inhibited by the hydroxamate-based metalloprotease inhibitors marimastat, TAPI-2, BB94, GM6001 and GW280264X, and by 10 nM of the tissue inhibitor of metalloproteinases (TIMP)-3, but not by up to 20 nM of TIMP-1 or -2. Additionally, we screened a non-hydroxamate small-molecule library for novel ADAM9 inhibitors and identified four compounds that selectively inhibited ADAM9-dependent proteolysis over ADAM10- or ADAM17-dependent processing. Taken together, the present study provides new information about the molecular fingerprint of ADAM9 in cell-based assays by showing that it is not stimulated by strong activators of ectodomain shedding and by defining a characteristic inhibitor profile. The identification of novel non-hydroxamate inhibitors of ADAM9 could provide the basis for designing more selective compounds that block the contribution of ADAM9 to pathological neovascularization and cancer.
Keywords: ADAM9 (A Disintegrin And Metalloproteinase 9); EphB4; ectodomain shedding; molecular fingerprint.
© 2017 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Figures


Similar articles
-
An Overview of ADAM9: Structure, Activation, and Regulation in Human Diseases.Int J Mol Sci. 2020 Oct 21;21(20):7790. doi: 10.3390/ijms21207790. Int J Mol Sci. 2020. PMID: 33096780 Free PMC article. Review.
-
Characterization of the catalytic activity of the membrane-anchored metalloproteinase ADAM15 in cell-based assays.Biochem J. 2009 Apr 28;420(1):105-13. doi: 10.1042/BJ20082127. Biochem J. 2009. PMID: 19207106
-
ADAM9 inhibition increases membrane activity of ADAM10 and controls α-secretase processing of amyloid precursor protein.J Biol Chem. 2011 Nov 25;286(47):40443-51. doi: 10.1074/jbc.M111.280495. Epub 2011 Sep 28. J Biol Chem. 2011. PMID: 21956108 Free PMC article.
-
ADAM10 is the major sheddase responsible for the release of membrane-associated meprin A.J Biol Chem. 2014 May 9;289(19):13308-22. doi: 10.1074/jbc.M114.559088. Epub 2014 Mar 24. J Biol Chem. 2014. PMID: 24662289 Free PMC article.
-
Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules.Comb Chem High Throughput Screen. 2005 Mar;8(2):161-71. doi: 10.2174/1386207053258488. Comb Chem High Throughput Screen. 2005. PMID: 15777180 Review.
Cited by
-
A Disintegrin and A Metalloproteinase-9 (ADAM9): A Novel Proteinase Culprit with Multifarious Contributions to COPD.Am J Respir Crit Care Med. 2018 Jun 4;198(12):1500-18. doi: 10.1164/rccm.201711-2300OC. Online ahead of print. Am J Respir Crit Care Med. 2018. PMID: 29864380 Free PMC article.
-
Sorting nexin 9 (SNX9) regulates levels of the transmembrane ADAM9 at the cell surface.J Biol Chem. 2018 May 25;293(21):8077-8088. doi: 10.1074/jbc.RA117.001077. Epub 2018 Apr 5. J Biol Chem. 2018. PMID: 29622675 Free PMC article.
-
ADAM9 promotes lung cancer progression through vascular remodeling by VEGFA, ANGPT2, and PLAT.Sci Rep. 2017 Nov 8;7(1):15108. doi: 10.1038/s41598-017-15159-1. Sci Rep. 2017. PMID: 29118335 Free PMC article.
-
An Overview of ADAM9: Structure, Activation, and Regulation in Human Diseases.Int J Mol Sci. 2020 Oct 21;21(20):7790. doi: 10.3390/ijms21207790. Int J Mol Sci. 2020. PMID: 33096780 Free PMC article. Review.
-
ADAM9 enhances Th17 cell differentiation and autoimmunity by activating TGF-β1.Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2023230118. doi: 10.1073/pnas.2023230118. Proc Natl Acad Sci U S A. 2021. PMID: 33911034 Free PMC article.
References
-
- Blobel CP (2005) ADAMs: key players in EGFR-signaling, development and disease. Nat. Rev. Mol. Cell. Bio. 6, 32–43 - PubMed
-
- Murphy G (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 12, 929–941 - PubMed
-
- Reiss K and Saftig P (2009) The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. 20, 126–137 - PubMed
-
- Weber S and Saftig P (2012) Ectodomain shedding and ADAMs in development. Development. 139, 3693–3709 - PubMed
-
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russel WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ and Black RA (1998) An essential role for ectodomain shedding in mammalian development. Science. 282,1281–1284 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

