Human Immunoglobulin Heavy Gamma Chain Polymorphisms: Molecular Confirmation Of Proteomic Assessment
- PMID: 28265047
- PMCID: PMC5417824
- DOI: 10.1074/mcp.M116.064733
Human Immunoglobulin Heavy Gamma Chain Polymorphisms: Molecular Confirmation Of Proteomic Assessment
Abstract
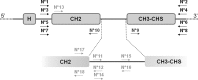
Immunoglobulin G (IgG) proteins are known for the huge diversity of the variable domains of their heavy and light chains, aimed at protecting each individual against foreign antigens. The IgG also harbor specific polymorphism concentrated in the CH2 and CH3-CHS constant regions located on the Fc fragment of their heavy chains. But this individual particularity relies only on a few amino acids among which some could make accurate sequence determination a challenge for mass spectrometry-based techniques.The purpose of the study was to bring a molecular validation of proteomic results by the sequencing of encoding DNA fragments. It was performed using ten individual samples (DNA and sera) selected on the basis of their Gm (gamma marker) allotype polymorphism in order to cover the main immunoglobulin heavy gamma (IGHG) gene diversity. Gm allotypes, reflecting part of this diversity, were determined by a serological method. On its side, the IGH locus comprises four functional IGHG genes totalizing 34 alleles and encoding the four IgG subclasses. The genomic study focused on the nucleotide polymorphism of the CH2 and CH3-CHS exons and of the intron. Despite strong sequence identity, four pairs of specific gene amplification primers could be designed. Additional primers were identified to perform the subsequent sequencing. The nucleotide sequences obtained were first assigned to a specific IGHG gene, and then IGHG alleles were deduced using a home-made decision tree reading of the nucleotide sequences. IGHG amino acid (AA) alleles were determined by mass spectrometry. Identical results were found at 95% between alleles identified by proteomics and those deduced from genomics. These results validate the proteomic approach which could be used for diagnostic purposes, namely for a mother-and-child differential IGHG detection in a context of suspicion of congenital infection.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures
References
-
- Lefranc M. P., and Lefranc G. (2001) The Immunoglobulin Factsbook, pp. 1–458, Academic Press, London, UK
-
- Lefranc M. P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., Hadi-Saljoqi S., Sasorith S., Lefranc G., and Kossida S. (2015) IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43, D413–D422 - PMC - PubMed
-
- Lefranc M. P., and Lefranc G. (2012) Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods Mol. Biol. 882, 635–680 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

