Selection of DNA aptamers with two modified bases
- PMID: 28265062
- PMCID: PMC5358403
- DOI: 10.1073/pnas.1615475114
Selection of DNA aptamers with two modified bases
Abstract
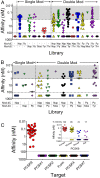
The nucleobases comprising DNA and RNA aptamers provide considerably less chemical diversity than protein-based ligands, limiting their versatility. The introduction of novel functional groups at just one of the four bases in modified aptamers has recently led to dramatic improvement in the success rate of identifying nucleic acid ligands to protein targets. Here we explore the benefits of additional enhancement in physicochemical diversity by selecting modified DNA aptamers that contain amino-acid-like modifications on both pyrimidine bases. Using proprotein convertase subtilisin/kexin type 9 as a representative protein target, we identify specific pairwise combinations of modifications that result in higher affinity, metabolic stability, and inhibitory potency compared with aptamers with single modifications. Such doubly modified aptamers are also more likely to be encoded in shorter sequences and occupy nonoverlapping epitopes more frequently than aptamers with single modifications. These highly modified DNA aptamers have broad utility in research, diagnostic, and therapeutic applications.
Keywords: PCSK9; PSMA; SELEX; SOMAmer; modified aptamer.
Conflict of interest statement
Conflict of interest statement: All authors are employees and/or shareholders of SomaLogic, Inc. SOMAmer reagent is a registered trademark of SomaLogic, Inc.
Figures



References
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. - PubMed
-
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. - PubMed
-
- Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31(5):453–457. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

