How conserved are the conserved 16S-rRNA regions?
- PMID: 28265511
- PMCID: PMC5333541
- DOI: 10.7717/peerj.3036
How conserved are the conserved 16S-rRNA regions?
Abstract
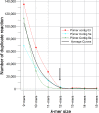
The 16S rRNA gene has been used as master key for studying prokaryotic diversity in almost every environment. Despite the claim of several researchers to have the best universal primers, the reality is that no primer has been demonstrated to be truly universal. This suggests that conserved regions of the gene may not be as conserved as expected. The aim of this study was to evaluate the conservation degree of the so-called conserved regions flanking the hypervariable regions of the 16S rRNA gene. Data contained in SILVA database (release 123) were used for the study. Primers reported as matches of each conserved region were assembled to form contigs; sequences sizing 12 nucleotides (12-mers) were extracted from these contigs and searched into the entire set of SILVA sequences. Frequency analysis shown that extreme regions, 1 and 10, registered the lowest frequencies. 12-mer frequencies revealed segments of contigs that were not as conserved as expected (≤90%). Fragments corresponding to the primer contigs 3, 4, 5b and 6a were recovered from all sequences in SILVA database. Nucleotide frequency analysis in each consensus demonstrated that only a small fraction of these so-called conserved regions is truly conserved in non-redundant sequences. It could be concluded that conserved regions of the 16S rRNA gene exhibit considerable variation that has to be considered when using this gene as biomarker.
Keywords: Biodiversity; Conserved regions 16S; Kmers; Primer design.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

Similar articles
-
An efficient strategy using k-mers to analyse 16S rRNA sequences.Heliyon. 2017 Jul 27;3(7):e00370. doi: 10.1016/j.heliyon.2017.e00370. eCollection 2017 Jul. Heliyon. 2017. PMID: 28795166 Free PMC article.
-
Characterization of the 18S rRNA gene for designing universal eukaryote specific primers.PLoS One. 2014 Feb 7;9(2):e87624. doi: 10.1371/journal.pone.0087624. eCollection 2014. PLoS One. 2014. PMID: 24516555 Free PMC article.
-
Primer Design for an Accurate View of Picocyanobacterial Community Structure by Using High-Throughput Sequencing.Appl Environ Microbiol. 2019 Mar 22;85(7):e02659-18. doi: 10.1128/AEM.02659-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30709827 Free PMC article.
-
Review and re-analysis of domain-specific 16S primers.J Microbiol Methods. 2003 Dec;55(3):541-55. doi: 10.1016/j.mimet.2003.08.009. J Microbiol Methods. 2003. PMID: 14607398 Review.
-
[Comparison between relative analogy and homology of 16S rRNA partial sequences between Mycobacterium szulgai and Mycobacterium malmoense].Kekkaku. 2003 Apr;78(4):359-63. Kekkaku. 2003. PMID: 12739396 Review. Japanese.
Cited by
-
Gut microbiome diversity detected by high-coverage 16S and shotgun sequencing of paired stool and colon sample.Sci Data. 2020 Mar 16;7(1):92. doi: 10.1038/s41597-020-0427-5. Sci Data. 2020. PMID: 32179734 Free PMC article.
-
Size-variable zone in V3 region of 16S rRNA.RNA Biol. 2017 Nov 2;14(11):1514-1521. doi: 10.1080/15476286.2017.1317912. Epub 2017 May 23. RNA Biol. 2017. PMID: 28440695 Free PMC article.
-
Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset.Microorganisms. 2024 Dec 25;13(1):16. doi: 10.3390/microorganisms13010016. Microorganisms. 2024. PMID: 39858784 Free PMC article.
-
An efficient strategy using k-mers to analyse 16S rRNA sequences.Heliyon. 2017 Jul 27;3(7):e00370. doi: 10.1016/j.heliyon.2017.e00370. eCollection 2017 Jul. Heliyon. 2017. PMID: 28795166 Free PMC article.
-
KnotAli: informed energy minimization through the use of evolutionary information.BMC Bioinformatics. 2022 May 3;23(1):159. doi: 10.1186/s12859-022-04673-3. BMC Bioinformatics. 2022. PMID: 35505276 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

