A supertree pipeline for summarizing phylogenetic and taxonomic information for millions of species
- PMID: 28265520
- PMCID: PMC5335690
- DOI: 10.7717/peerj.3058
A supertree pipeline for summarizing phylogenetic and taxonomic information for millions of species
Abstract
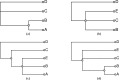
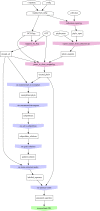
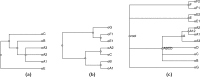
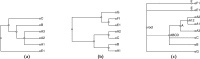
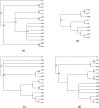
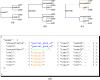
We present a new supertree method that enables rapid estimation of a summary tree on the scale of millions of leaves. This supertree method summarizes a collection of input phylogenies and an input taxonomy. We introduce formal goals and criteria for such a supertree to satisfy in order to transparently and justifiably represent the input trees. In addition to producing a supertree, our method computes annotations that describe which grouping in the input trees support and conflict with each group in the supertree. We compare our supertree construction method to a previously published supertree construction method by assessing their performance on input trees used to construct the Open Tree of Life version 4, and find that our method increases the number of displayed input splits from 35,518 to 39,639 and decreases the number of conflicting input splits from 2,760 to 1,357. The new supertree method also improves on the previous supertree construction method in that it produces no unsupported branches and avoids unnecessary polytomies. This pipeline is currently used by the Open Tree of Life project to produce all of the versions of project's "synthetic tree" starting at version 5. This software pipeline is called "propinquity". It relies heavily on "otcetera"-a set of C++ tools to perform most of the steps of the pipeline. All of the components are free software and are available on GitHub.
Keywords: Phylogenetics; Software; Supertree; Taxonomy; Tree of life.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



References
-
- Aho AV, Sagiv Y, Szymanski TG, Ullman JD. Inferring a tree from lowest common ancestors with an application to the optimization of relational expressions. SIAM Journal on Computing. 1981;10(3):405–421. doi: 10.1137/0210030. - DOI
-
- Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41(1):3–10. doi: 10.2307/1222480. - DOI
-
- Gatesy J, Springer MS. A critique of matrix representation with parsimony supertrees. In: Bininda-Edmonds ORP, editor. Phylogenetic supertrees: combining information to reveal the tree of life. vol. 3. Dordrecht: Springer; 2004. pp. 369–388. (Computational biology). Dress A, ed.
LinkOut - more resources
Full Text Sources
Other Literature Sources

