MiR-9 is involved in TGF-β1-induced lung cancer cell invasion and adhesion by targeting SOX7
- PMID: 28266181
- PMCID: PMC5571535
- DOI: 10.1111/jcmm.13120
MiR-9 is involved in TGF-β1-induced lung cancer cell invasion and adhesion by targeting SOX7
Abstract
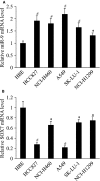
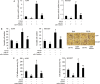
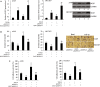
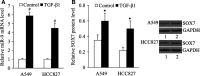
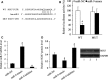
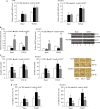
MicroRNA (miR)-9 plays different roles in different cancer types. Here, we investigated the role of miR-9 in non-small-cell lung cancer (NSCLC) cell invasion and adhesion in vitro and explored whether miR-9 was involved in transforming growth factor-beta 1 (TGF-β1)-induced NSCLC cell invasion and adhesion by targeting SOX7. The expression of miR-9 and SOX7 in human NSCLC tissues and cell lines was examined by reverse transcription-quantitative polymerase chain reaction. Gain-of-function and loss-of-function experiments were performed on A549 and HCC827 cells to investigate the effect of miR-9 and SOX7 on NSCLC cell invasion and adhesion in the presence or absence of TGF-β1. Transwell-Matrigel assay and cell adhesion assay were used to examine cell invasion and adhesion abilities. Luciferase reporter assay was performed to determine whether SOX7 was a direct target of miR-9. We found miR-9 was up-regulated and SOX7 was down-regulated in human NSCLC tissues and cell lines. Moreover, SOX7 expression was negatively correlated with miR-9 expression. miR-9 knockdown or SOX7 overexpression could suppress TGF-β1-induced NSCLC cell invasion and adhesion. miR-9 directly targets the 3' untranslated region of SOX7, and SOX7 protein expression was down-regulated by miR-9. TGF-β1 induced miR-9 expression in NSCLC cells. miR-9 up-regulation led to enhanced NSCLC cell invasion and adhesion; however, these effects could be attenuated by SOX7 overexpression. We concluded that miR-9 expression was negatively correlated with SOX7 expression in human NSCLC. miR-9 was up-regulated by TGF-β1 and contributed to TGF-β1-induced NSCLC cell invasion and adhesion by directly targeting SOX7.
Keywords: SOX7; adhesion; invasion; lung cancer; microRNA-9; transforming growth factor-beta 1.
© 2017 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures
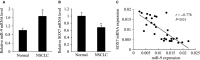
Similar articles
-
MicroRNA-616 promotes the growth and metastasis of non-small cell lung cancer by targeting SOX7.Oncol Rep. 2017 Oct;38(4):2078-2086. doi: 10.3892/or.2017.5854. Epub 2017 Jul 28. Oncol Rep. 2017. Retraction in: Oncol Rep. 2023 Oct;50(4):186. doi: 10.3892/or.2023.8623. PMID: 28765960 Free PMC article. Retracted.
-
MiR-145 and miR-203 represses TGF-β-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells.Lung Cancer. 2016 Jul;97:87-94. doi: 10.1016/j.lungcan.2016.04.017. Epub 2016 Apr 27. Lung Cancer. 2016. PMID: 27237033
-
MicroRNA-494-3p Promotes Cell Growth, Migration, and Invasion of Nasopharyngeal Carcinoma by Targeting Sox7.Technol Cancer Res Treat. 2018 Jan 1;17:1533033818809993. doi: 10.1177/1533033818809993. Technol Cancer Res Treat. 2018. PMID: 30381030 Free PMC article.
-
SOX7: from a developmental regulator to an emerging tumor suppressor.Histol Histopathol. 2014 Apr;29(4):439-45. doi: 10.14670/HH-29.10.439. Epub 2013 Nov 29. Histol Histopathol. 2014. PMID: 24288056 Free PMC article. Review.
-
Novel role of sex-determining region Y-box 7 (SOX7) in tumor biology and cardiovascular developmental biology.Semin Cancer Biol. 2020 Dec;67(Pt 1):49-56. doi: 10.1016/j.semcancer.2019.08.032. Epub 2019 Aug 29. Semin Cancer Biol. 2020. PMID: 31473269 Review.
Cited by
-
A Likely Role for a Novel Cell Therapeutic Target of Transforming Growth Factor-β1 on Radiation Pneumonitis in Lung and Nasopharyngeal Cancer Patients.Cell Transplant. 2020 Jan-Dec;29:963689720914245. doi: 10.1177/0963689720914245. Cell Transplant. 2020. PMID: 32252552 Free PMC article.
-
Salvianolic acid A improves nerve regeneration and repairs nerve defects in rats with brain injury by downregulating miR-212-3p-mediated SOX7.Kaohsiung J Med Sci. 2023 Dec;39(12):1222-1232. doi: 10.1002/kjm2.12779. Epub 2023 Nov 21. Kaohsiung J Med Sci. 2023. PMID: 37987200 Free PMC article.
-
The role of cancer-derived microRNAs in cancer immune escape.J Hematol Oncol. 2020 Mar 28;13(1):25. doi: 10.1186/s13045-020-00848-8. J Hematol Oncol. 2020. PMID: 32222150 Free PMC article. Review.
-
LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis.Onco Targets Ther. 2017 Oct 25;10:5137-5149. doi: 10.2147/OTT.S146423. eCollection 2017. Onco Targets Ther. 2017. PMID: 29123412 Free PMC article.
-
Zfx-induced upregulation of UBE2J1 facilitates endometrial cancer progression via PI3K/AKT pathway.Cancer Biol Ther. 2021 Mar 4;22(3):238-247. doi: 10.1080/15384047.2021.1883186. Epub 2021 Feb 26. Cancer Biol Ther. 2021. PMID: 33632059 Free PMC article.
References
-
- Jemal A, Siegel R, Xu J, et al Cancer statistics, 2010. CA Cancer J Clin. 2010; 60: 277–300. - PubMed
-
- Jemal A, Siegel R, Ward E, et al Cancer statistics, 2008. CA Cancer J Clin. 2008; 58: 71–96. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell. 1993; 75: 843–54. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin‐14 by lin‐4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75: 855–62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

