Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus
- PMID: 28266557
- PMCID: PMC5339811
- DOI: 10.1038/srep43733
Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus
Abstract
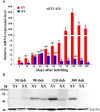
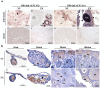
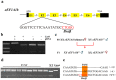
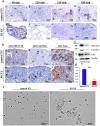
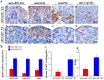
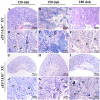
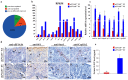
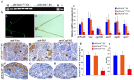
Eukaryotic elongation factor 1 alpha (eEF1A) is an essential component of the translational apparatus. In the present study, eEF1A1b was isolated from the Nile tilapia. Real-time PCR and Western blot revealed that eEF1A1b was expressed highly in the testis from 90 dah (days after hatching) onwards. In situ hybridization and immunohistochemistry analyses showed that eEF1A1b was highly expressed in the spermatogonia of the testis. CRISPR/Cas9 mediated mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in the F0 XY fish. Consistently, heterozygous mutation of eEF1A1b (eEF1A1b+/-) resulted in an absence of spermatocytes at 90 dah, very few spermatocytes, spermatids and spermatozoa at 180 dah, and decreased Cyp11b2 and serum 11-ketotestosterone level at both stages. Further examination of the fertilization capacity of the sperm indicated that the eEF1A1b+/- XY fish were infertile due to abnormal spermiogenesis. Transcriptomic analyses of the eEF1A1b+/- testis from 180 dah XY fish revealed that key elements involved in spermatogenesis, steroidogenesis and sperm motility were significantly down-regulated compared with the control XY. Transgenic overexpression of eEF1A1b rescued the spermatogenesis arrest phenotype of the eEF1A1b+/- testis. Taken together, our data suggested that eEF1A1b is crucial for spermatogenesis and male fertility in the Nile tilapia.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia.Cell Mol Life Sci. 2020 Dec;77(23):4921-4938. doi: 10.1007/s00018-019-03439-0. Epub 2020 Jan 18. Cell Mol Life Sci. 2020. PMID: 31955242 Free PMC article.
-
The role of StAR2 gene in testicular differentiation and spermatogenesis in Nile tilapia (Oreochromis niloticus).J Steroid Biochem Mol Biol. 2021 Nov;214:105974. doi: 10.1016/j.jsbmb.2021.105974. Epub 2021 Aug 20. J Steroid Biochem Mol Biol. 2021. PMID: 34425195
-
Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia.Biol Reprod. 2014 Dec;91(6):136. doi: 10.1095/biolreprod.114.121418. Epub 2014 Oct 15. Biol Reprod. 2014. PMID: 25320148
-
Haploid male germ cells-the Grand Central Station of protein transport.Hum Reprod Update. 2020 Jun 18;26(4):474-500. doi: 10.1093/humupd/dmaa004. Hum Reprod Update. 2020. PMID: 32318721 Review.
-
Spermatogenesis in mammals: proteomic insights.Syst Biol Reprod Med. 2012 Aug;58(4):179-90. doi: 10.3109/19396368.2012.691943. Syst Biol Reprod Med. 2012. PMID: 22788530 Review.
Cited by
-
Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia.Cell Mol Life Sci. 2020 Dec;77(23):4921-4938. doi: 10.1007/s00018-019-03439-0. Epub 2020 Jan 18. Cell Mol Life Sci. 2020. PMID: 31955242 Free PMC article.
-
Sustainable use of CRISPR/Cas in fish aquaculture: the biosafety perspective.Transgenic Res. 2022 Feb;31(1):1-21. doi: 10.1007/s11248-021-00274-7. Epub 2021 Jul 25. Transgenic Res. 2022. PMID: 34304349 Free PMC article. Review.
-
Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay.DNA Res. 2019 Aug 1;26(4):313-325. doi: 10.1093/dnares/dsz011. DNA Res. 2019. PMID: 31173071 Free PMC article.
-
Genome editing in East African cichlids and tilapias: state-of-the-art and future directions.Open Biol. 2023 Nov;13(11):230257. doi: 10.1098/rsob.230257. Epub 2023 Nov 29. Open Biol. 2023. PMID: 38018094 Free PMC article. Review.
-
Physiological impact and comparison of mutant screening methods in piwil2 KO founder Nile tilapia produced by CRISPR/Cas9 system.Sci Rep. 2020 Jul 28;10(1):12600. doi: 10.1038/s41598-020-69421-0. Sci Rep. 2020. PMID: 32724054 Free PMC article.
References
-
- Koji N., Masataka K., Shigekazu N. & Yoshito K. Structure of the two genes coding for polypeptide chain elongation factor lα (EF-lα) froni Saccharomyces cerevisiae. Gene 45, 265–273 (1986). - PubMed
-
- Danforth B. N. & Ji S. Elongation factor-1 alpha occurs as two copies in bees: implications for phylogenetic analysis of EF-1 alpha sequences in insects. Molecular biology and evolution 15, 225–235 (1998). - PubMed
-
- Gao D., Li Z., Murphy T. & Sauerbier W. Structure and transcription of the gene for translation elongation factor 1 subunit alpha of zebrafish (Danio rerio). Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1350, 1–5 (1997). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

