Commensal gut bacteria modulate phosphorylation-dependent PPARγ transcriptional activity in human intestinal epithelial cells
- PMID: 28266623
- PMCID: PMC5339702
- DOI: 10.1038/srep43199
Commensal gut bacteria modulate phosphorylation-dependent PPARγ transcriptional activity in human intestinal epithelial cells
Abstract
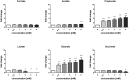
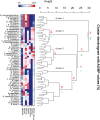
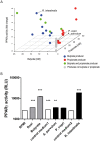
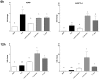
In healthy subjects, the intestinal microbiota interacts with the host's epithelium, regulating gene expression to the benefit of both, host and microbiota. The underlying mechanisms remain poorly understood, however. Although many gut bacteria are not yet cultured, constantly growing culture collections have been established. We selected 57 representative commensal bacterial strains to study bacteria-host interactions, focusing on PPARγ, a key nuclear receptor in colonocytes linking metabolism and inflammation to the microbiota. Conditioned media (CM) were harvested from anaerobic cultures and assessed for their ability to modulate PPARγ using a reporter cell line. Activation of PPARγ transcriptional activity was linked to the presence of butyrate and propionate, two of the main metabolites of intestinal bacteria. Interestingly, some stimulatory CMs were devoid of these metabolites. A Prevotella and an Atopobium strain were chosen for further study, and shown to up-regulate two PPARγ-target genes, ANGPTL4 and ADRP. The molecular mechanisms of these activations involved the phosphorylation of PPARγ through ERK1/2. The responsible metabolites were shown to be heat sensitive but markedly diverged in size, emphasizing the diversity of bioactive compounds found in the intestine. Here we describe different mechanisms by which single intestinal bacteria can directly impact their host's health through transcriptional regulation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






Similar articles
-
Commensal Streptococcus salivarius Modulates PPARγ Transcriptional Activity in Human Intestinal Epithelial Cells.PLoS One. 2015 May 6;10(5):e0125371. doi: 10.1371/journal.pone.0125371. eCollection 2015. PLoS One. 2015. PMID: 25946041 Free PMC article.
-
Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells.PLoS One. 2012;7(12):e52869. doi: 10.1371/journal.pone.0052869. Epub 2012 Dec 27. PLoS One. 2012. PMID: 23300800 Free PMC article.
-
Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids.mBio. 2014 Aug 12;5(4):e01438-14. doi: 10.1128/mBio.01438-14. mBio. 2014. PMID: 25118238 Free PMC article.
-
The intestinal epithelium as guardian of gut barrier integrity.Cell Microbiol. 2015 Nov;17(11):1561-9. doi: 10.1111/cmi.12501. Epub 2015 Sep 15. Cell Microbiol. 2015. PMID: 26294173 Review.
-
PPARγ: The Central Mucus Barrier Coordinator in Ulcerative Colitis.Inflamm Bowel Dis. 2021 Apr 15;27(5):732-741. doi: 10.1093/ibd/izaa273. Inflamm Bowel Dis. 2021. PMID: 33772551 Review.
Cited by
-
The Crosstalk between Microbiome and Mitochondrial Homeostasis in Neurodegeneration.Cells. 2023 Jan 28;12(3):429. doi: 10.3390/cells12030429. Cells. 2023. PMID: 36766772 Free PMC article. Review.
-
Intertwined Relationship of Mitochondrial Metabolism, Gut Microbiome and Exercise Potential.Int J Mol Sci. 2022 Feb 28;23(5):2679. doi: 10.3390/ijms23052679. Int J Mol Sci. 2022. PMID: 35269818 Free PMC article. Review.
-
Nuclear receptors: a bridge linking the gut microbiome and the host.Mol Med. 2021 Nov 5;27(1):144. doi: 10.1186/s10020-021-00407-y. Mol Med. 2021. PMID: 34740314 Free PMC article. Review.
-
Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns.PeerJ. 2020 May 22;8:e9205. doi: 10.7717/peerj.9205. eCollection 2020. PeerJ. 2020. PMID: 32509465 Free PMC article.
-
Regulation of Oxygen Homeostasis at the Intestinal Epithelial Barrier Site.Int J Mol Sci. 2021 Aug 25;22(17):9170. doi: 10.3390/ijms22179170. Int J Mol Sci. 2021. PMID: 34502078 Free PMC article. Review.
References
-
- Sekirov I., Russell S. L., Antunes L. C. M. & Finlay B. B. Gut Microbiota in Health and Disease. Physiol. Rev. 90, 859–904 (2010). - PubMed
-
- Cerf-Bensussan N. & Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 10, 735–44 (2010). - PubMed
-
- Cummings J. H. Microbial Digestion of Complex Carbohydrates in Man. Proc. Nutr. Soc. 43, 35–44 (1984). - PubMed
-
- Albert M. J., Mathan V. I. & Baker S. J. Vitamin B12 synthesis by human small intestinal bacteria. Nature 283, 781–782 (1980). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

