Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability
- PMID: 28268090
- PMCID: PMC5412515
- DOI: 10.1016/j.dnarep.2017.02.012
Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability
Abstract
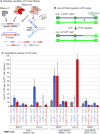
Cells carrying deletions of genes encoding H-class ribonucleases display elevated rates of chromosome instability. The role of these enzymes is to remove RNA-DNA associations including persistent mRNA-DNA hybrids (R-loops) formed during transcription, and ribonucleotides incorporated into DNA during replication. RNases H1 and H2 can degrade the RNA component of R-loops, but only RNase H2 can initiate accurate ribonucleotide excision repair (RER). In order to examine the specific contributions of these activities to chromosome stability, we measured rates of loss-of-heterozygosity (LOH) in diploid Saccharomyces cerevisiae yeast strains carrying the rnh201-RED separation-of-function allele, encoding a version of RNase H2 that is RER-defective, but partly retains its other activity. The LOH rate in rnh201-RED was intermediate between RNH201 and rnh201Δ. In strains carrying a mutant version of DNA polymerase ε (pol2-M644G) that incorporates more ribonucleotides than normal, the LOH rate in rnh201-RED was as high as the rate measured in rnh201Δ. Topoisomerase 1 cleavage at sites of ribonucleotide incorporation has been recently shown to produce DNA double strand breaks. Accordingly, in both the POL2 and pol2-M644G backgrounds, the LOH elevation in rnh201-RED was suppressed by top1Δ. In contrast, in strains that incorporate fewer ribonucleotides (pol2-M644L) the LOH rate in rnh201-RED was low and independent of topoisomerase 1. These results suggest that both R-loop removal and RER contribute substantially to chromosome stability, and that their relative contributions may be variable across different regions of the genome. In this scenario, a prominent contribution of R-loop removal may be expected at highly transcribed regions, whereas RER may play a greater role at hotspots of ribonucleotide incorporation.
Keywords: Loss-of-heterozygosity (LOH); R-loops; RNH201; RNase H2; Ribonucleotides.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
-
- Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nature Reviews Genetics. 2015;16:583–597. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

