A framework for informing segmentation of in vivo MRI with information derived from ex vivo imaging: Application in the medial temporal lobe
- PMID: 28269623
- PMCID: PMC5603287
- DOI: 10.1109/EMBC.2016.7592099
A framework for informing segmentation of in vivo MRI with information derived from ex vivo imaging: Application in the medial temporal lobe
Abstract
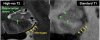
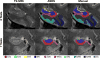
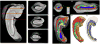
Automatic segmentation of cortical and subcortical structures is commonplace in brain MRI literature and is frequently used as the first step towards quantitative analysis of structural and functional neuroimaging. Most approaches to brain structure segmentation are based on propagation of anatomical information from example MRI datasets, called atlases or templates, that are manually labeled by experts. The accuracy of automatic segmentation is usually validated against the "bronze" standard of manual segmentation of test MRI datasets. However, good performance vis-a-vis manual segmentation does not imply accuracy relative to the underlying true anatomical boundaries. In the context of segmentation of hippocampal subfields and functionally related medial temporal lobe cortical subregions, we explore the challenges associated with validating existing automatic segmentation techniques against underlying histologically-derived anatomical "gold" standard; and, further, developing automatic in vivo MRI segmentation techniques informed by histological imaging.
Figures



Similar articles
-
Clinical Application of Automatic Segmentation of Medial Temporal Lobe Subregions in Prodromal and Dementia-Level Alzheimer's Disease.J Alzheimers Dis. 2016 Oct 4;54(3):1027-1037. doi: 10.3233/JAD-160014. J Alzheimers Dis. 2016. PMID: 27567809 Free PMC article.
-
Structural and functional asymmetry of medial temporal subregions in unilateral temporal lobe epilepsy: A 7T MRI study.Hum Brain Mapp. 2019 Jun 1;40(8):2390-2398. doi: 10.1002/hbm.24530. Epub 2019 Jan 21. Hum Brain Mapp. 2019. PMID: 30666753 Free PMC article.
-
Development of a histologically validated segmentation protocol for the hippocampal body.Neuroimage. 2017 Aug 15;157:219-232. doi: 10.1016/j.neuroimage.2017.06.008. Epub 2017 Jun 3. Neuroimage. 2017. PMID: 28587896
-
MRI segmentation of the human brain: challenges, methods, and applications.Comput Math Methods Med. 2015;2015:450341. doi: 10.1155/2015/450341. Epub 2015 Mar 1. Comput Math Methods Med. 2015. PMID: 25945121 Free PMC article. Review.
-
Neonatal brain MRI segmentation: A review.Comput Biol Med. 2015 Sep;64:163-78. doi: 10.1016/j.compbiomed.2015.06.016. Epub 2015 Jun 29. Comput Biol Med. 2015. PMID: 26189155 Review.
Cited by
-
Precise MRI-histology coregistration of paraffin-embedded tissue with blockface imaging.Imaging Neurosci (Camb). 2025 Aug 8;3:IMAG.a.106. doi: 10.1162/IMAG.a.106. eCollection 2025. Imaging Neurosci (Camb). 2025. PMID: 40800888 Free PMC article.
-
Tensor image registration library: Deformable registration of stand-alone histology images to whole-brain post-mortem MRI data.Neuroimage. 2023 Jan;265:119792. doi: 10.1016/j.neuroimage.2022.119792. Epub 2022 Dec 9. Neuroimage. 2023. PMID: 36509214 Free PMC article.
References
-
- de Flores R, La Joie R, Chételat G. Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience. 2015 Nov;309:29–50. - PubMed
-
- Yushkevich PA, Amaral RSC, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, Chételat G, Daugherty AM, Davachi L, Ding S-L, Ekstrom A, Geerlings MI, Hassan A, Huang Y, Iglesias JE, La Joie R, Kerchner GA, LaRocque KF, Libby LA, Malykhin N, Mueller SG, Olsen RK, Palombo DJ, Parekh MB, Pluta JB, Preston AR, Pruessner JC, Ranganath C, Raz N, Schlichting ML, Schoemaker D, Singh S, Stark CEL, Suthana N, Tompary A, Turowski MM, Van Leemput K, Wagner AD, Wang L, Winterburn JL, Wisse LEM, Yassa MA, Zeineh MM, for the Hippocampal Subfields Group (HSG) Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage. 2015 Jan; - PMC - PubMed
-
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K, Alzheimer’s Disease Neuroimaging Initiative A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage. 2015 Jul;115:117–37. - PMC - PubMed
-
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding S-L, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Human Brain Mapping. 2014 [Online]. Available: http://dx.doi.org/10.1002/hbm.22627. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
