Microevolution of Serial Clinical Isolates of Cryptococcus neoformans var. grubii and C. gattii
- PMID: 28270580
- PMCID: PMC5340869
- DOI: 10.1128/mBio.00166-17
Microevolution of Serial Clinical Isolates of Cryptococcus neoformans var. grubii and C. gattii
Abstract
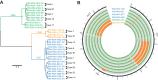
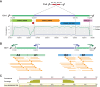
The pathogenic species of Cryptococcus are a major cause of mortality owing to severe infections in immunocompromised as well as immunocompetent individuals. Although antifungal treatment is usually effective, many patients relapse after treatment, and in such cases, comparative analyses of the genomes of incident and relapse isolates may reveal evidence of determinative, microevolutionary changes within the host. Here, we analyzed serial isolates cultured from cerebrospinal fluid specimens of 18 South African patients with recurrent cryptococcal meningitis. The time between collection of the incident isolates and collection of the relapse isolates ranged from 124 days to 290 days, and the analyses revealed that, during this period within the patients, the isolates underwent several genetic and phenotypic changes. Considering the vast genetic diversity of cryptococcal isolates in sub-Saharan Africa, it was not surprising to find that the relapse isolates had acquired different genetic and correlative phenotypic changes. They exhibited various mechanisms for enhancing virulence, such as growth at 39°C, adaptation to stress, and capsule production; a remarkable amplification of ERG11 at the native and unlinked locus may provide stable resistance to fluconazole. Our data provide a deeper understanding of the microevolution of Cryptococcus species under pressure from antifungal chemotherapy and host immune responses. This investigation clearly suggests a promising strategy to identify novel targets for improved diagnosis, therapy, and prognosis.IMPORTANCE Opportunistic infections caused by species of the pathogenic yeast Cryptococcus lead to chronic meningoencephalitis and continue to ravage thousands of patients with HIV/AIDS. Despite receiving antifungal treatment, over 10% of patients develop recurrent disease. In this study, we collected isolates of Cryptococcus from cerebrospinal fluid specimens of 18 patients at the time of their diagnosis and when they relapsed several months later. We then sequenced and compared the genomic DNAs of each pair of initial and relapse isolates. We also tested the isolates for several key properties related to cryptococcal virulence as well as for their susceptibility to the antifungal drug fluconazole. These analyses revealed that the relapsing isolates manifested multiple genetic and chromosomal changes that affected a variety of genes implicated in the pathogenicity of Cryptococcus or resistance to fluconazole. This application of comparative genomics to serial clinical isolates provides a blueprint for identifying the mechanisms whereby pathogenic microbes adapt within patients to prolong disease.
Copyright © 2017 Chen et al.
Figures


References
-
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101:17258–17263. doi:10.1073/pnas.0402981101. - DOI - PMC - PubMed
-
- Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA; Cryptococcus gattii Working Group of the Pacific Northwest . 2009. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis 15:1185–1191. doi:10.3201/eid1508.081384. - DOI - PMC - PubMed
-
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon CC, Billmyre RB, Brunel F, Bahn YS, Chen W, Chen Y, Chow EWL, Coppée JY, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh YP, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, et al. . 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. doi:10.1371/journal.pgen.1004261. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
