Viruses in the Oceanic Basement
- PMID: 28270584
- PMCID: PMC5340873
- DOI: 10.1128/mBio.02129-16
Viruses in the Oceanic Basement
Abstract
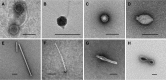
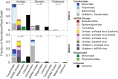
Microbial life has been detected well into the igneous crust of the seafloor (i.e., the oceanic basement), but there have been no reports confirming the presence of viruses in this habitat. To detect and characterize an ocean basement virome, geothermally heated fluid samples (ca. 60 to 65°C) were collected from 117 to 292 m deep into the ocean basement using seafloor observatories installed in two boreholes (Integrated Ocean Drilling Program [IODP] U1362A and U1362B) drilled in the eastern sediment-covered flank of the Juan de Fuca Ridge. Concentrations of virus-like particles in the fluid samples were on the order of 0.2 × 105 to 2 × 105 ml-1 (n = 8), higher than prokaryote-like cells in the same samples by a factor of 9 on average (range, 1.5 to 27). Electron microscopy revealed diverse viral morphotypes similar to those of viruses known to infect bacteria and thermophilic archaea. An analysis of virus-like sequences in basement microbial metagenomes suggests that those from archaeon-infecting viruses were the most common (63 to 80%). Complete genomes of a putative archaeon-infecting virus and a prophage within an archaeal scaffold were identified among the assembled sequences, and sequence analysis suggests that they represent lineages divergent from known thermophilic viruses. Of the clustered regularly interspaced short palindromic repeat (CRISPR)-containing scaffolds in the metagenomes for which a taxonomy could be inferred (163 out of 737), 51 to 55% appeared to be archaeal and 45 to 49% appeared to be bacterial. These results imply that the warmed, highly altered fluids in deeply buried ocean basement harbor a distinct assemblage of novel viruses, including many that infect archaea, and that these viruses are active participants in the ecology of the basement microbiome.IMPORTANCE The hydrothermally active ocean basement is voluminous and likely provided conditions critical to the origins of life, but the microbiology of this vast habitat is not well understood. Viruses in particular, although integral to the origins, evolution, and ecology of all life on earth, have never been documented in basement fluids. This report provides the first estimate of free virus particles (virions) within fluids circulating through the extrusive basalt of the seafloor and describes the morphological and genetic signatures of basement viruses. These data push the known geographical limits of the virosphere deep into the ocean basement and point to a wealth of novel viral diversity, exploration of which could shed light on the early evolution of viruses.
Copyright © 2017 Nigro et al.
Figures




References
-
- Kimura M, Jia Z-J, Nakayama N, Asakawa S. 2008. Ecology of viruses in soils: past, present and future perspectives. Soil Sci Plant Nutr 54:1–32. doi: 10.1111/j.1747-0765.2007.00197.x. - DOI
-
- Wilhelm SW, Matteson AR. 2008. Freshwater and marine virioplankton: a brief overview of commonalities and differences. Freshw Biol 53:1076–1089. doi: 10.1111/j.1365-2427.2008.01980.x. - DOI
-
- Danovaro R, Corinaldesi C, Filippini M, Fischer UR, Gessner MO, Jacquet S, Magagnini M, Velimirov B. 2008. Viriobenthos in freshwater and marine sediments: a review. Freshw Biol 53:1186–1213. doi: 10.1111/j.1365-2427.2008.01961.x. - DOI
-
- Bird DF, Juniper SK, Ricciardi-Rigault M, Martineu P, Prairie YT, Calvert SE. 2001. Subsurface viruses and bacteria in Holocene-Late Pleistocene sediments of Saanich Inlet, BC: ODP Holes 1033B and 1034B, Leg 169S. Mar Geol 174:227–239. doi: 10.1016/S0025-3227(00)00152-3. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
