DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons
- PMID: 28270689
- PMCID: PMC5344974
- DOI: 10.1038/ncomms14712
DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons
Erratum in
-
Author Correction: DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons.Nat Commun. 2020 Sep 14;11(1):4696. doi: 10.1038/s41467-020-18562-x. Nat Commun. 2020. PMID: 32929092 Free PMC article.
Abstract
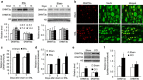
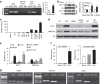
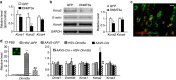
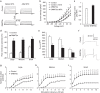
Nerve injury induces changes in gene transcription in dorsal root ganglion (DRG) neurons, which may contribute to nerve injury-induced neuropathic pain. DNA methylation represses gene expression. Here, we report that peripheral nerve injury increases expression of the DNA methyltransferase DNMT3a in the injured DRG neurons via the activation of the transcription factor octamer transcription factor 1. Blocking this increase prevents nerve injury-induced methylation of the voltage-dependent potassium (Kv) channel subunit Kcna2 promoter region and rescues Kcna2 expression in the injured DRG and attenuates neuropathic pain. Conversely, in the absence of nerve injury, mimicking this increase reduces the Kcna2 promoter activity, diminishes Kcna2 expression, decreases Kv current, increases excitability in DRG neurons and leads to spinal cord central sensitization and neuropathic pain symptoms. These findings suggest that DNMT3a may contribute to neuropathic pain by repressing Kcna2 expression in the DRG.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Mitka M. Virtual textbook on pain developed: effort seeks to remedy gap in medical education. J. Am. Med. Assoc. 290, 2395 (2003). - PubMed
-
- Chung J. M. & Chung K. Importance of hyperexcitability of DRG neurons in neuropathic pain. Pain Pract. 2, 87–97 (2002). - PubMed
-
- Devor M. Ectopic discharge in Abeta afferents as a source of neuropathic pain. Exp. Brain Res. 196, 115–128 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

