Understanding Epstein-Barr Virus Life Cycle with Proteomics: A Temporal Analysis of Ubiquitination During Virus Reactivation
- PMID: 28271981
- PMCID: PMC5240003
- DOI: 10.1089/omi.2016.0158
Understanding Epstein-Barr Virus Life Cycle with Proteomics: A Temporal Analysis of Ubiquitination During Virus Reactivation
Abstract
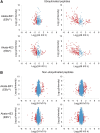
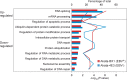
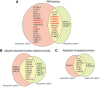
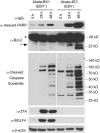
Epstein-Barr virus (EBV) is a human γ-herpesvirus associated with cancer, including Burkitt lymphoma, nasopharyngeal, and gastric carcinoma. EBV reactivation in latently infected B cells is essential for persistent infection whereby B cell receptor (BCR) activation is a physiologically relevant stimulus. Yet, a global view of BCR activation-regulated protein ubiquitination is lacking when EBV is actively replicating. We report here, for the first time, the long-term effects of IgG cross-linking-regulated protein ubiquitination and offer a basis for dissecting the cellular environment during the course of EBV lytic replication. Using the Akata-BX1 (EBV+) and Akata-4E3 (EBV-) Burkitt lymphoma cells, we monitored the dynamic changes in protein ubiquitination using quantitative proteomics. We observed temporal alterations in the level of ubiquitination at ∼150 sites in both EBV+ and EBV- B cells post-IgG cross-linking, compared with controls with no cross-linking. The majority of protein ubiquitination was downregulated. The upregulated ubiquitination events were associated with proteins involved in RNA processing. Among the downregulated ubiquitination events were proteins involved in apoptosis, ubiquitination, and DNA repair. These comparative and quantitative proteomic observations represent the first analysis on the effects of IgG cross-linking at later time points when the majority of EBV genes are expressed and the viral genome is actively being replicated. In all, these data enhance our understanding of mechanistic linkages connecting protein ubiquitination, RNA processing, apoptosis, and the EBV life cycle.
Keywords: Association Study; big data; proteomics.
Conflict of interest statement
The authors declare that no conflicting financial interests exist.
Figures



References
-
- Adhikari A, and Chen ZJ. (2009). Diversity of polyubiquitin chains. Dev Cell 16, 485–486 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

