Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae
- PMID: 28272414
- PMCID: PMC5344976
- DOI: 10.1038/ncomms14665
Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae
Abstract
DNA-binding proteins are central regulators of chromosome organization; however, in genome-reduced bacteria their diversity is largely diminished. Whether the chromosomes of such bacteria adopt defined three-dimensional structures remains unexplored. Here we combine Hi-C and super-resolution microscopy to determine the structure of the Mycoplasma pneumoniae chromosome at a 10 kb resolution. We find a defined structure, with a global symmetry between two arms that connect opposite poles, one bearing the chromosomal Ori and the other the midpoint. Analysis of local structures at a 3 kb resolution indicates that the chromosome is organized into domains ranging from 15 to 33 kb. We provide evidence that genes within the same domain tend to be co-regulated, suggesting that chromosome organization influences transcriptional regulation, and that supercoiling regulates local organization. This study extends the current understanding of bacterial genome organization and demonstrates that a defined chromosomal structure is a universal feature of living systems.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

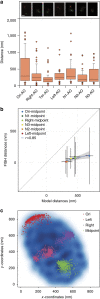
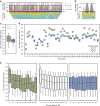
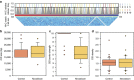

Similar articles
-
Structure and function of the bacterial chromosome.Trends Biochem Sci. 1988 Apr;13(4):131-5. doi: 10.1016/0968-0004(88)90069-2. Trends Biochem Sci. 1988. PMID: 3075377 Review. No abstract available.
-
Architectural organization in E. coli nucleoid.Biochim Biophys Acta. 2012 Jul;1819(7):830-5. doi: 10.1016/j.bbagrm.2012.02.012. Epub 2012 Feb 22. Biochim Biophys Acta. 2012. PMID: 22387214 Free PMC article. Review.
-
High-resolution 3D models of Caulobacter crescentus chromosome reveal genome structural variability and organization.Nucleic Acids Res. 2018 May 4;46(8):3937-3952. doi: 10.1093/nar/gky141. Nucleic Acids Res. 2018. PMID: 29529244 Free PMC article.
-
Chromosomal domains of supercoiling in Salmonella typhimurium.Mol Microbiol. 1993 Nov;10(3):685-96. doi: 10.1111/j.1365-2958.1993.tb00940.x. Mol Microbiol. 1993. PMID: 7968545
-
High-resolution mapping of the spatial organization of a bacterial chromosome.Science. 2013 Nov 8;342(6159):731-4. doi: 10.1126/science.1242059. Epub 2013 Oct 24. Science. 2013. PMID: 24158908 Free PMC article.
Cited by
-
A Hi-C data-integrated model elucidates E. coli chromosome's multiscale organization at various replication stages.Nucleic Acids Res. 2021 Apr 6;49(6):3077-3091. doi: 10.1093/nar/gkab094. Nucleic Acids Res. 2021. PMID: 33660781 Free PMC article.
-
Protein cleavage influences surface protein presentation in Mycoplasma pneumoniae.Sci Rep. 2021 Mar 24;11(1):6743. doi: 10.1038/s41598-021-86217-y. Sci Rep. 2021. PMID: 33762641 Free PMC article.
-
Bacterial live therapeutics for human diseases.Mol Syst Biol. 2024 Dec;20(12):1261-1281. doi: 10.1038/s44320-024-00067-0. Epub 2024 Oct 23. Mol Syst Biol. 2024. PMID: 39443745 Free PMC article. Review.
-
Single-nucleotide-resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome.Nucleic Acids Res. 2019 Feb 20;47(3):1373-1388. doi: 10.1093/nar/gky1222. Nucleic Acids Res. 2019. PMID: 30517674 Free PMC article.
-
Rules and Exceptions: The Role of Chromosomal ParB in DNA Segregation and Other Cellular Processes.Microorganisms. 2020 Jan 11;8(1):0. doi: 10.3390/microorganisms8010105. Microorganisms. 2020. PMID: 31940850 Free PMC article. Review.
References
-
- Cremer T. & Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001). - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007). - PubMed
-
- Dorman C. J. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11, 349–355 (2013). - PubMed
-
- Dillon S. C. & Dorman C. J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

