Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle
- PMID: 28272430
- PMCID: PMC5341059
- DOI: 10.1038/srep44026
Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle
Abstract
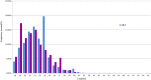
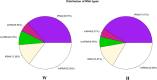
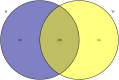
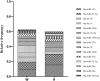
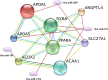
MicroRNAs (miRNAs) are important post-transcriptional regulators involved in animal adipogenesis, however, their roles in bovine fat deposition remain poorly understood. In the present study, we conducted a comparative RNA sequencing to identify the key miRNAs involved in beef lipid accumulation by comparing the backfat small RNA samples between Wagyu (high intramuscular fat) and Holstein (moderate intramuscular fat) cattle. Fifteen miRNAs such as bta-miR-142-3p, bta-miR-379, bta-miR-196a, bta-miR-196b, bta-miR-30f and bta-miR-2887 were identified to have a higher expression level in Wagyu cattle compared with Holstein, whereas bta-miR-320a, bta-miR-874 and bta-miR-1247-3p had a lower expression level in Wagyu. Furthermore, a total of 1345 potential target genes of differentially expressed miRNAs were predicted using bioinformatics tools, in which PPARα and RXRα were known to play a critical role in adipocyte differentiation and lipid metabolism. In conclusion, the present study constructed a high-throughput RNA sequencing screen and successfully identified miRNAs such as bta-miR-874, bta-miR-320a and bta-miR-196b which may affect beef fat deposition. The present findings may provide a theoretical foundation for the utilization of beef cattle germplasm resources.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

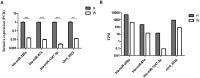
References
-
- Owens F. N., Dubeski P. & Hanson C. F. Factors that alter the growth and development of ruminants. J.Anim.Sci. 71, 3138–3150 (1993). - PubMed
-
- Large V., Peroni O., Letexier D., Ray H. & Beylot M. Metabolism of lipids in human white adipocyte. Diabetes & Metab. 30, 294–309 (2004). - PubMed
-
- Hausman G. J., Basu U., Wei S., Hausman D. B. & Dodson M. V. Preadipocyte and adipose tissue differentiation in meat animals: influence of species and anatomical location. Annu. Rev. Anim. Biosci. 2, 323–351 (2014). - PubMed
-
- Miao X. & Luo Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 145, 587–596 (2013). - PubMed
-
- Miao X., Luo Q. & Qin X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene 582, 69–76 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

