Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources
- PMID: 28272476
- PMCID: PMC5341023
- DOI: 10.1038/srep43797
Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources
Abstract
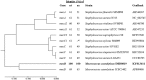
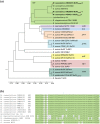
Methicillin-resistant Macrococcus caseolyticus strains from bovine and canine origins were found to carry a novel mecD gene conferring resistance to all classes of β-lactams including anti-MRSA cephalosporins. Association of β-lactam resistance with mecD was demonstrated by gene expression in S. aureus and deletion of the mecD-containing island in M. caseolyticus. The mecD gene was located either on an 18,134-bp M. caseolyticus resistance island (McRImecD-1) or a 16,188-bp McRImecD-2. Both islands were integrated at the 3' end of the rpsI gene, carried the mecD operon (mecD-mecR1m-mecIm), and genes for an integrase of the tyrosine recombinase family and a putative virulence-associated protein (virE). Apart from the mecD operon, that shared 66% overall nucleotide identity with the mecB operon, McRImecD islands were unrelated to any mecB-carrying elements or staphylococcal cassette chromosome mec. Only McRImecD-1 that is delimitated at both ends by direct repeats was capable of circular excision. The recombined excision pattern suggests site-specific activity of the integrase and allowed identification of a putative core attachment site. Detection of rpsI-associated integrases in Bacillus and S. aureus reveals a potential for broad-host range dissemination of the novel methicillin resistance gene mecD.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Kloos W. E. et al.. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipericus sp. nov., and Macrococcus bovicus sp. no. and Macrococcus carouselicus sp. nov. Int. J. Syst. Bacteriol. 48 Pt 3, 859–877, doi: 10.1099/00207713-48-3-859 (1998). - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

