Mechanism-Based Inhibition of the Mycobacterium tuberculosis Branched-Chain Aminotransferase by d- and l-Cycloserine
- PMID: 28272868
- PMCID: PMC5834943
- DOI: 10.1021/acschembio.7b00142
Mechanism-Based Inhibition of the Mycobacterium tuberculosis Branched-Chain Aminotransferase by d- and l-Cycloserine
Abstract
The branched-chain aminotransferase is a pyridoxal 5'-phosphate (PLP)-dependent enzyme responsible for the final step in the biosynthesis of all three branched-chain amino acids, l-leucine, l-isoleucine, and l-valine, in bacteria. We have investigated the mechanism of inactivation of the branched-chain aminotransferase from Mycobacterium tuberculosis (MtIlvE) by d- and l-cycloserine. d-Cycloserine is currently used only in the treatment of multidrug-drug-resistant tuberculosis. Our results show a time- and concentration-dependent inactivation of MtIlvE by both isomers, with l-cycloserine being a 40-fold better inhibitor of the enzyme. Minimum inhibitory concentration (MIC) studies revealed that l-cycloserine is a 10-fold better inhibitor of Mycobacterium tuberculosis growth than d-cycloserine. In addition, we have crystallized the MtIlvE-d-cycloserine inhibited enzyme, determining the structure to 1.7 Å. The structure of the covalent d-cycloserine-PMP adduct bound to MtIlvE reveals that the d-cycloserine ring is planar and aromatic, as previously observed for other enzyme systems. Mass spectrometry reveals that both the d-cycloserine- and l-cycloserine-PMP complexes have the same mass, and are likely to be the same aromatized, isoxazole product. However, the kinetics of formation of the MtIlvE d-cycloserine-PMP and MtIlvE l-cycloserine-PMP adducts are quite different. While the kinetics of the formation of the MtIlvE d-cycloserine-PMP complex can be fit to a single exponential, the formation of the MtIlvE l-cycloserine-PMP complex occurs in two steps. We propose a chemical mechanism for the inactivation of d- and l-cycloserine which suggests a stereochemically determined structural role for the differing kinetics of inactivation. These results demonstrate that the mechanism of action of d-cycloserine's activity against M. tuberculosis may be more complicated than previously thought and that d-cycloserine may compromise the in vivo activity of multiple PLP-dependent enzymes, including MtIlvE.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
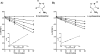


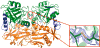

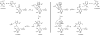
Similar articles
-
Chemical Mechanism of the Branched-Chain Aminotransferase IlvE from Mycobacterium tuberculosis.Biochemistry. 2016 Nov 15;55(45):6295-6303. doi: 10.1021/acs.biochem.6b00928. Epub 2016 Nov 2. Biochemistry. 2016. PMID: 27780341 Free PMC article.
-
Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation.Mol Biosyst. 2010 Sep;6(9):1682-93. doi: 10.1039/c003743e. Epub 2010 May 5. Mol Biosyst. 2010. PMID: 20445930 Free PMC article.
-
The 1.9 A structure of the branched-chain amino-acid transaminase (IlvE) from Mycobacterium tuberculosis.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Nov 1;65(Pt 11):1071-7. doi: 10.1107/S1744309109036690. Epub 2009 Oct 13. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19923721 Free PMC article.
-
Inhibition of 7,8-diaminopelargonic acid aminotransferase by amiclenomycin and analogues.Biochem Soc Trans. 2005 Aug;33(Pt 4):802-5. doi: 10.1042/BST0330802. Biochem Soc Trans. 2005. PMID: 16042602 Review.
-
Molecular basis underlying Mycobacterium tuberculosis D-cycloserine resistance. Is there a role for ubiquinone and menaquinone metabolic pathways?Expert Opin Ther Targets. 2014 Jun;18(6):691-701. doi: 10.1517/14728222.2014.902937. Epub 2014 Apr 29. Expert Opin Ther Targets. 2014. PMID: 24773568 Review.
Cited by
-
Use of Human Lung Tissue Models for Screening of Drugs against SARS-CoV-2 Infection.Viruses. 2022 Oct 31;14(11):2417. doi: 10.3390/v14112417. Viruses. 2022. PMID: 36366514 Free PMC article.
-
The mechanism of branched-chain amino acid transferases in different diseases: Research progress and future prospects.Front Oncol. 2022 Sep 2;12:988290. doi: 10.3389/fonc.2022.988290. eCollection 2022. Front Oncol. 2022. PMID: 36119495 Free PMC article. Review.
-
Bacterial Branched-Chain Amino Acid Biosynthesis: Structures, Mechanisms, and Drugability.Biochemistry. 2017 Nov 7;56(44):5849-5865. doi: 10.1021/acs.biochem.7b00849. Biochemistry. 2017. PMID: 28977745 Free PMC article. Review.
-
PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.Chem Commun (Camb). 2019 Nov 28;55(96):14502-14505. doi: 10.1039/c9cc06975e. Chem Commun (Camb). 2019. PMID: 31730149 Free PMC article.
-
Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.ACS Chem Biol. 2019 Apr 19;14(4):742-750. doi: 10.1021/acschembio.9b00030. Epub 2019 Mar 13. ACS Chem Biol. 2019. PMID: 30830751 Free PMC article.
References
-
- World Health Organization releases. 2015 global report on tuberculosis. Breathe. 2015;11:244–244.
-
- Franco TMA, Rostirolla DC, Ducati RG, Lorenzini DM, Basso LA, Santos DS. Biochemical characterization of recombinant guaA-encoded guanosine monophosphate synthetase (EC 6.3.5.2) from Mycobacterium tuberculosis H37Rv strain. Arch Biochem Biophys. 2012;517:1–11. - PubMed
-
- Neuhaus FC. Antibiotics I, Mechanism of Action. Springer, Verlag; New York: 1967. pp. 40–83.
-
- Strominger J, Ito E, Threnn RH. Competitive Inhibition of Enzymatic Reactions by Oxamycin. J Am Chem Soc. 1960;82:998–999.
-
- Soper TS, Manning JM. Different modes of action of inhibitors of bacterial D-amino acid transaminase. A target enzyme for the design of new antibacterial agents. J Biol Chem. 1981;256:4263–4268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

