Hypersensitive termination of the hypoxic response by a disordered protein switch
- PMID: 28273070
- PMCID: PMC5375031
- DOI: 10.1038/nature21705
Hypersensitive termination of the hypoxic response by a disordered protein switch
Abstract
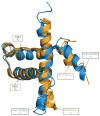
The cellular response to hypoxia is critical for cell survival and is fine-tuned to allow cells to recover from hypoxic stress and adapt to heterogeneous or fluctuating oxygen levels. The hypoxic response is mediated by the α-subunit of the transcription factor HIF-1 (HIF-1α), which interacts through its intrinsically disordered C-terminal transactivation domain with the TAZ1 (also known as CH1) domain of the general transcriptional coactivators CBP and p300 to control the transcription of critical adaptive genes. One such gene encodes CITED2, a negative feedback regulator that attenuates HIF-1 transcriptional activity by competing for TAZ1 binding through its own disordered transactivation domain. Little is known about the molecular mechanism by which CITED2 displaces the tightly bound HIF-1α from their common cellular target. The HIF-1α and CITED2 transactivation domains bind to TAZ1 through helical motifs that flank a conserved LP(Q/E)L sequence that is essential for negative feedback regulation. Here we show that human CITED2 displaces HIF-1α by forming a transient ternary complex with TAZ1 and HIF-1α and competing for a shared binding site through its LPEL motif, thus promoting a conformational change in TAZ1 that increases the rate of HIF-1α dissociation. Through allosteric enhancement of HIF-1α release, CITED2 activates a highly responsive negative feedback circuit that rapidly and efficiently attenuates the hypoxic response, even at modest CITED2 concentrations. This hypersensitive regulatory switch is entirely dependent on the unique flexibility and binding properties of these intrinsically disordered proteins and probably exemplifies a common strategy used by the cell to respond rapidly to environmental signals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Comment in
-
Molecular biology: A hidden competitive advantage of disorder.Nature. 2017 Mar 16;543(7645):325-326. doi: 10.1038/nature21893. Epub 2017 Mar 8. Nature. 2017. PMID: 28273062 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

