Immune protection against reinfection with nonprimate hepacivirus
- PMID: 28275093
- PMCID: PMC5373355
- DOI: 10.1073/pnas.1619380114
Immune protection against reinfection with nonprimate hepacivirus
Abstract
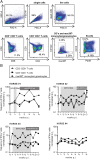
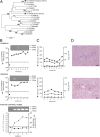
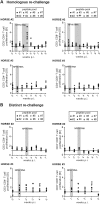
Hepatitis C virus (HCV) displays a restricted host species tropism and only humans and chimpanzees are susceptible to infection. A robust immunocompetent animal model is still lacking, hampering mechanistic analysis of virus pathogenesis, immune control, and prophylactic vaccine development. The closest homolog of HCV is the equine nonprimate hepacivirus (NPHV), which shares similar features with HCV and thus represents an animal model to study hepacivirus infections in their natural hosts. We aimed to dissect equine immune responses after experimental NPHV infection and conducted challenge experiments to investigate immune protection against secondary NPHV infections. Horses were i.v. injected with NPHV containing plasma. Flow cytometric analysis was used to monitor immune cell frequencies and activation status. All infected horses became viremic after 1 or 2 wk and viremia could be detected in two horses for several weeks followed by a delayed seroconversion and viral clearance. Histopathological examinations of liver biopsies revealed mild, periportally accentuated infiltrations of lymphocytes, macrophages, and plasma cells with some horses displaying subclinical signs of hepatitis. Following viral challenge, an activation of equine immune responses was observed. Importantly, after a primary NPHV infection, horses were protected against rechallenge with the homologous as well as a distinct isolate with only minute amounts of circulating virus being detectable.
Keywords: hepatitis C virus; immune protection; infection; nonprimate hepacivirus; rechallenge.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Frequent occurrence of nonprimate hepacivirus infections in Thoroughbred breeding horses - A cross-sectional study for the occurrence of infections and potential risk factors.Vet Microbiol. 2017 May;203:315-322. doi: 10.1016/j.vetmic.2017.03.030. Epub 2017 Apr 4. Vet Microbiol. 2017. PMID: 28619163
-
Prevalence of Equine Hepacivirus Infections in France and Evidence for Two Viral Subtypes Circulating Worldwide.Transbound Emerg Dis. 2017 Dec;64(6):1884-1897. doi: 10.1111/tbed.12587. Epub 2016 Nov 24. Transbound Emerg Dis. 2017. PMID: 27882682
-
Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses.Hepatology. 2015 Feb;61(2):447-59. doi: 10.1002/hep.27440. Epub 2015 Jan 5. Hepatology. 2015. PMID: 25212983
-
What Do We Know About Hepatitis Viruses in Horses?Vet Clin North Am Equine Pract. 2019 Aug;35(2):351-362. doi: 10.1016/j.cveq.2019.03.001. Epub 2019 May 10. Vet Clin North Am Equine Pract. 2019. PMID: 31084975 Review.
-
Surveying the global virome: identification and characterization of HCV-related animal hepaciviruses.Antiviral Res. 2015 Mar;115:83-93. doi: 10.1016/j.antiviral.2014.12.014. Epub 2014 Dec 26. Antiviral Res. 2015. PMID: 25545071 Free PMC article. Review.
Cited by
-
Viral testing of 10 cases of Theiler's disease and 37 in-contact horses in the absence of equine biologic product administration: A prospective study (2014-2018).J Vet Intern Med. 2019 Jan;33(1):258-265. doi: 10.1111/jvim.15362. Epub 2018 Dec 6. J Vet Intern Med. 2019. PMID: 30520132 Free PMC article.
-
Hepacivirus A Infection in Horses Defines Distinct Envelope Hypervariable Regions and Elucidates Potential Roles of Viral Strain and Adaptive Immune Status in Determining Envelope Diversity and Infection Outcome.J Virol. 2018 Aug 29;92(18):e00314-18. doi: 10.1128/JVI.00314-18. Print 2018 Sep 15. J Virol. 2018. Retraction in: J Virol. 2020 Dec 9;95(1):e01963-20. doi: 10.1128/JVI.01963-20. PMID: 29976666 Free PMC article. Retracted.
-
Pathogenesis, MicroRNA-122 Gene-Regulation, and Protective Immune Responses After Acute Equine Hepacivirus Infection.Hepatology. 2021 Sep;74(3):1148-1163. doi: 10.1002/hep.31802. Epub 2021 Jun 11. Hepatology. 2021. PMID: 33713356 Free PMC article.
-
Further Evidence for in Utero Transmission of Equine Hepacivirus to Foals.Viruses. 2019 Dec 5;11(12):1124. doi: 10.3390/v11121124. Viruses. 2019. PMID: 31817371 Free PMC article.
-
Hepatitis C Virus Vaccine Research: Time to Put Up or Shut Up.Viruses. 2021 Aug 12;13(8):1596. doi: 10.3390/v13081596. Viruses. 2021. PMID: 34452460 Free PMC article. Review.
References
-
- WHO 2016 Hepatitis C: Fact Sheet no. 164. Available at www.who.int/mediacentre/factsheets/fs164/en/. Accessed February 28, 2017.
-
- Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10(9):553–562. - PubMed
-
- Simmonds P. Genetic diversity and evolution of hepatitis C virus: 15 years on. J Gen Virol. 2004;85(Pt 11):3173–3188. - PubMed
-
- Reddy KR, et al. All-oral direct-acting antiviral therapy in HCV-advanced liver disease is effective in real-world practice: Observations through HCV-TARGET database. Alimentary Pharmacology and Therapeutics. 2017;45(1):115–126. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

