DNA supercoiling during transcription
- PMID: 28275417
- PMCID: PMC5338639
- DOI: 10.1007/s12551-016-0215-9
DNA supercoiling during transcription
Abstract
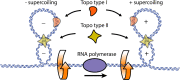
The twin-supercoiled-domain model describes how transcription can drive DNA supercoiling, and how DNA supercoiling, in turn plays an important role in regulating gene transcription. In vivo and in vitro experiments have disclosed many details of the complex interactions in this relationship, and recently new insights have been gained with the help of genome-wide DNA supercoiling mapping techniques and single molecule methods. This review summarizes the general mechanisms of the interplay between DNA supercoiling and transcription, considers the biological implications, and focuses on recent important discoveries and technical advances in this field. We highlight the significant impact of DNA supercoiling in transcription, but also more broadly in all processes operating on DNA.
Keywords: chromatin; gene regulation; mechanics; supercoiling; torque; transcription.
Conflict of interest statement
Conflict of interest
None.
Ethical approval
This article does not contain any studies with human or animal subjects performed by the authors.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

