SIDT2 mediates gymnosis, the uptake of naked single-stranded oligonucleotides into living cells
- PMID: 28277980
- PMCID: PMC5785214
- DOI: 10.1080/15476286.2017.1302641
SIDT2 mediates gymnosis, the uptake of naked single-stranded oligonucleotides into living cells
Abstract
Single-stranded oligonucleotides (ssOligos) are efficiently taken up by living cells without the use of transfection reagents. This phenomenon called 'gymnosis' enables the sequence-specific silencing of target genes in various types of cells. Several antisense ssOligos are used for the treatment of human diseases. However, the molecular mechanism underlying the uptake of naked ssOligos into cells remains to be elucidated. Here, we show that systemic RNA interference deficient-1 (SID-1) transmembrane family 2 (SIDT2), a mammalian ortholog of the Caenorhabditis elegans double-stranded RNA channel SID-1, mediates gymnosis. We show that the uptake of naked ssOligos into cells is significantly downregulated by knockdown of SIDT2. Furthermore, knockdown of SIDT2 inhibited the effect of antisense RNA mediated by gymnosis. Overexpression of SIDT2 enhanced the uptake of naked ssOligos into cells, while a single amino acid mutation in SIDT2 abolished this effect. Our findings highlight the mechanism of extra- and intracellular RNA transport and may contribute to the further development of nucleic acid-based therapies.
Keywords: Gymnosis; RNA channel; SIDT2; membrane protein; single-stranded oligonucleotides.
Figures
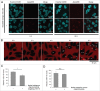
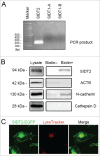
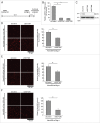
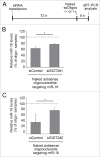

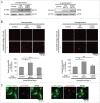
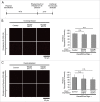
References
-
- Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nat Rev Drug Discov 2002; 1:503-14; PMID:12120257; https://doi.org/ 10.1038/nrd837 - DOI - PubMed
-
- Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A 1984; 81:1966-70; PMID:6201848; https://doi.org/ 10.1073/pnas.81.7.1966 - DOI - PMC - PubMed
-
- Neves MA, Reinstein O, Saad M, Johnson PE. Defining the secondary structural requirements of a cocaine-binding aptamer by a thermodynamic and mutation study. Biophys Chem 2010; 153:9-16; PMID:21035241; https://doi.org/ 10.1016/j.bpc.2010.09.009 - DOI - PubMed
-
- Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J Mol Biol 2000; 301:117-28; PMID:10926496; https://doi.org/ 10.1006/jmbi.2000.3951 - DOI - PubMed
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; https://doi.org/ 10.1038/35888 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
