Mismatch repair proteins recruited to ultraviolet light-damaged sites lead to degradation of licensing factor Cdt1 in the G1 phase
- PMID: 28278049
- PMCID: PMC5397274
- DOI: 10.1080/15384101.2017.1295179
Mismatch repair proteins recruited to ultraviolet light-damaged sites lead to degradation of licensing factor Cdt1 in the G1 phase
Abstract
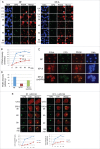
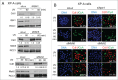
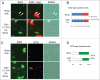
Cdt1 is rapidly degraded by CRL4Cdt2 E3 ubiquitin ligase after UV (UV) irradiation. Previous reports revealed that the nucleotide excision repair (NER) pathway is responsible for the rapid Cdt1-proteolysis. Here, we show that mismatch repair (MMR) proteins are also involved in the degradation of Cdt1 after UV irradiation in the G1 phase. First, compared with the rapid (within ∼15 min) degradation of Cdt1 in normal fibroblasts, Cdt1 remained stable for ∼30 min in NER-deficient XP-A cells, but was degraded within ∼60 min. The delayed degradation was also dependent on PCNA and CRL4Cdt2. The MMR proteins Msh2 and Msh6 were recruited to the UV-damaged sites of XP-A cells in the G1 phase. Depletion of these factors with small interfering RNAs prevented Cdt1 degradation in XP-A cells. Similar to the findings in XP-A cells, depletion of XPA delayed Cdt1 degradation in normal fibroblasts and U2OS cells, and co-depletion of Msh6 further prevented Cdt1 degradation. Furthermore, depletion of Msh6 alone delayed Cdt1 degradation in both cell types. When Cdt1 degradation was attenuated by high Cdt1 expression, repair synthesis at the damaged sites was inhibited. Our findings demonstrate that UV irradiation induces multiple repair pathways that activate CRL4Cdt2 to degrade its target proteins in the G1 phase of the cell cycle, leading to efficient repair of DNA damage.
Keywords: CRL4Cdt2; Cdt1; Mismatch repair (MMR); PCNA; UV.
Figures



Comment in
-
Mismatch repair regulates Cdt1 after UV damage.Cell Cycle. 2017 Jun 18;16(12):1143-1144. doi: 10.1080/15384101.2017.1319687. Cell Cycle. 2017. PMID: 28426347 Free PMC article. No abstract available.
Similar articles
-
Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells.J Biol Chem. 2010 Dec 31;285(53):41993-2000. doi: 10.1074/jbc.M110.161661. Epub 2010 Oct 7. J Biol Chem. 2010. PMID: 20929861 Free PMC article.
-
Two different replication factor C proteins, Ctf18 and RFC1, separately control PCNA-CRL4Cdt2-mediated Cdt1 proteolysis during S phase and following UV irradiation.Mol Cell Biol. 2012 Jun;32(12):2279-88. doi: 10.1128/MCB.06506-11. Epub 2012 Apr 9. Mol Cell Biol. 2012. PMID: 22493068 Free PMC article.
-
Checkpoint kinase ATR phosphorylates Cdt2, a substrate receptor of CRL4 ubiquitin ligase, and promotes the degradation of Cdt1 following UV irradiation.PLoS One. 2012;7(9):e46480. doi: 10.1371/journal.pone.0046480. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029527 Free PMC article.
-
Regulation of DNA Replication Licensing and Re-Replication by Cdt1.Int J Mol Sci. 2021 May 14;22(10):5195. doi: 10.3390/ijms22105195. Int J Mol Sci. 2021. PMID: 34068957 Free PMC article. Review.
-
The Licensing Factor Cdt1 Links Cell Cycle Progression to the DNA Damage Response.Anticancer Res. 2020 May;40(5):2449-2456. doi: 10.21873/anticanres.14214. Anticancer Res. 2020. PMID: 32366388 Review.
Cited by
-
Mismatch repair regulates Cdt1 after UV damage.Cell Cycle. 2017 Jun 18;16(12):1143-1144. doi: 10.1080/15384101.2017.1319687. Cell Cycle. 2017. PMID: 28426347 Free PMC article. No abstract available.
-
Cellular Responses to Platinum-Based Anticancer Drugs and UVC: Role of p53 and Implications for Cancer Therapy.Int J Mol Sci. 2020 Aug 11;21(16):5766. doi: 10.3390/ijms21165766. Int J Mol Sci. 2020. PMID: 32796711 Free PMC article. Review.
-
Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2 E3 ligase activity to Cdt1.Life Sci Alliance. 2018 Dec 31;1(6):e201800238. doi: 10.26508/lsa.201800238. eCollection 2018 Dec. Life Sci Alliance. 2018. PMID: 30623174 Free PMC article.
-
Genome-Wide DNA Alterations in X-Irradiated Human Gingiva Fibroblasts.Int J Mol Sci. 2020 Aug 12;21(16):5778. doi: 10.3390/ijms21165778. Int J Mol Sci. 2020. PMID: 32806598 Free PMC article.
-
STK19 is a DNA/RNA-binding protein critical for DNA damage repair and cell proliferation.J Cell Biol. 2024 Feb 5;223(2):e202301090. doi: 10.1083/jcb.202301090. Epub 2024 Jan 22. J Cell Biol. 2024. PMID: 38252411 Free PMC article.
References
-
- Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell 2010; 40:179-204; PMID:20965415; http://dx.doi.org/10.1016/j.molcel.2010.09.019 - DOI - PMC - PubMed
-
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: Where, when, and how? Annu Rev Biochem 2010; 79:89-130; PMID:20373915; http://dx.doi.org/10.1146/annurev.biochem.052308.103205 - DOI - PubMed
-
- Havens CG, Walter JC. Mechanism of CRL4Cdt2, a PCNA-dependent E3 ubiquitin ligase. Genes Dev 2011; 25:1568-82; PMID:21828267; http://dx.doi.org/10.1101/gad.2068611 - DOI - PMC - PubMed
-
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709-21; PMID:16949367; http://dx.doi.org/10.1016/j.molcel.2006.08.010 - DOI - PubMed
-
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev 2006; 20:3117-29; PMID:17085480; http://dx.doi.org/10.1101/gad.1482106 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
