Suppression of the grainyhead transcription factor 2 gene (GRHL2) inhibits the proliferation, migration, invasion and mediates cell cycle arrest of ovarian cancer cells
- PMID: 28278050
- PMCID: PMC5397264
- DOI: 10.1080/15384101.2017.1295181
Suppression of the grainyhead transcription factor 2 gene (GRHL2) inhibits the proliferation, migration, invasion and mediates cell cycle arrest of ovarian cancer cells
Abstract
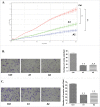
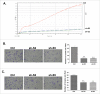
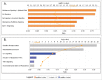
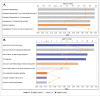
Previously, we have identified the Grainyhead transcription factor 2 gene (GRHL2) as notably hypomethylated in high-grade (HG) serous epithelial ovarian tumors, compared with normal ovarian tissues. GRHL2 is known for its functions in normal tissue development and wound healing. In the context of cancer, the role of GRHL2 is still ambiguous as both tumorigenic and tumor suppressive functions have been reported for this gene, although a role of GRHL2 in maintaining the epithelial status of cancer cells has been suggested. In this study, we report that GRHL2 is strongly overexpressed in both low malignant potential (LMP) and HG serous epithelial ovarian tumors, which probably correlates with its hypomethylated status. Suppression of the GRHL2 expression led to a sharp decrease in cell proliferation, migration and invasion and induced G1 cell cycle arrest in epithelial ovarian cancer (EOC) cells displaying either epithelial (A2780s) or mesenchymal (SKOV3) phenotypes. However, no phenotypic alterations were observed in these EOC cell lines following GRHL2 silencing. Gene expression profiling and consecutive canonical pathway and network analyses confirmed these data, as in both these EOC cell lines, GRHL2 ablation was associated with the downregulation of various genes and pathways implicated in cell growth and proliferation, cell cycle control and cellular metabolism. Taken together, our data are indicative for a strong oncogenic potential of the GRHL2 gene in EOC progression and support recent findings on the role of GRHL2 as one of the major phenotypic stability factors (PSFs) that stabilize the highly aggressive/metastatic hybrid epithelial/mesenchymal (E/M) phenotype of cancer cells.
Keywords: CRISPR/Cas9; GRHL2; cancer cell migration/invasion; epithelial ovarian cancer; epithelial-to-mesenchymal transition; microarrays; shRNA.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5-29; PMID:25559415; http://dx.doi.org/10.3322/caac.21254 - DOI - PubMed
-
- Marchetti C, Pisano C, Facchini G, Bruni GS, Magazzino FP, Losito S, Pignata S. First-line treatment of advanced ovarian cancer: Current research and perspectives. Expert Rev Anticancer Ther 2010; 10:47-60; PMID:20014885; http://dx.doi.org/10.1586/era.09.167 - DOI - PubMed
-
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683-92; PMID:17320506; http://dx.doi.org/10.1016/j.cell.2007.01.029 - DOI - PMC - PubMed
-
- Balch C, Fang F, Matei DE, Huang TH, Nephew KP. Minireview: Epigenetic changes in ovarian cancer. Endocrinology 2009; 150:4003-11; PMID:19574400; http://dx.doi.org/10.1210/en.2009-0404 - DOI - PMC - PubMed
-
- Bauerschlag DO, Ammerpohl O, Brautigam K, Schem C, Lin Q, Weigel MT, Hilpert F, Arnold N, Maass N, Meinhold-Heerlein I, et al.. Progression-free survival in ovarian cancer is reflected in epigenetic DNA methylation profiles. Oncology 2011; 80:12-20; PMID:21577013; http://dx.doi.org/10.1159/000327746 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
