Validation of diffusion tensor MRI measurements of cardiac microstructure with structure tensor synchrotron radiation imaging
- PMID: 28279178
- PMCID: PMC5345150
- DOI: 10.1186/s12968-017-0342-x
Validation of diffusion tensor MRI measurements of cardiac microstructure with structure tensor synchrotron radiation imaging
Abstract
Background: Diffusion tensor imaging (DTI) is widely used to assess tissue microstructure non-invasively. Cardiac DTI enables inference of cell and sheetlet orientations, which are altered under pathological conditions. However, DTI is affected by many factors, therefore robust validation is critical. Existing histological validation is intrinsically flawed, since it requires further tissue processing leading to sample distortion, is routinely limited in field-of-view and requires reconstruction of three-dimensional volumes from two-dimensional images. In contrast, synchrotron radiation imaging (SRI) data enables imaging of the heart in 3D without further preparation following DTI. The objective of the study was to validate DTI measurements based on structure tensor analysis of SRI data.
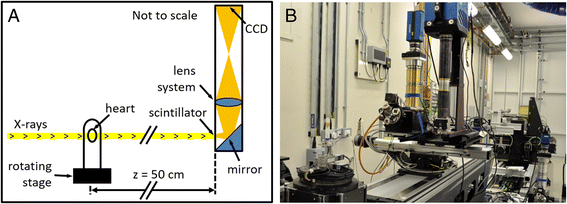
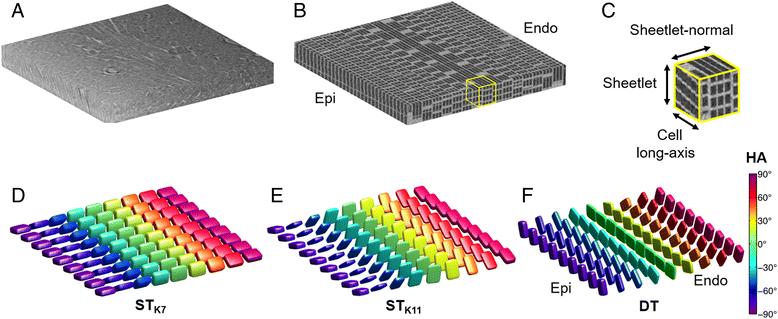
Methods: One isolated, fixed rat heart was imaged ex vivo with DTI and X-ray phase contrast SRI, and reconstructed at 100 μm and 3.6 μm isotropic resolution respectively. Structure tensors were determined from the SRI data and registered to the DTI data.
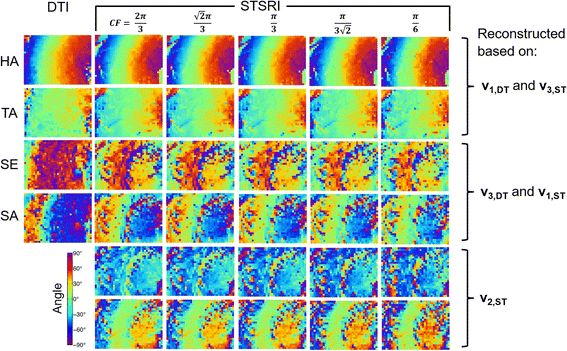
Results: Excellent agreement in helix angles (HA) and transverse angles (TA) was observed between the DTI and structure tensor synchrotron radiation imaging (STSRI) data, where HADTI-STSRI = -1.4° ± 23.2° and TADTI-STSRI = -1.4° ± 35.0° (mean ± 1.96 standard deviation across all voxels in the left ventricle). STSRI confirmed that the primary eigenvector of the diffusion tensor corresponds with the cardiomyocyte long-axis across the whole myocardium.
Conclusions: We have used STSRI as a novel and high-resolution gold standard for the validation of DTI, allowing like-with-like comparison of three-dimensional tissue structures in the same intact heart free of distortion. This represents a critical step forward in independently verifying the structural basis and informing the interpretation of cardiac DTI data, thereby supporting the further development and adoption of DTI in structure-based electro-mechanical modelling and routine clinical applications.
Keywords: Cardiovascular magnetic resonance; Diffusion tensor imaging; Structure tensor; Synchrotron radiation imaging; Tissue microstructure; Validation.
Figures






Similar articles
-
Comparison of diffusion tensor imaging by cardiovascular magnetic resonance and gadolinium enhanced 3D image intensity approaches to investigation of structural anisotropy in explanted rat hearts.J Cardiovasc Magn Reson. 2015 Apr 29;17(1):31. doi: 10.1186/s12968-015-0129-x. J Cardiovasc Magn Reson. 2015. PMID: 25926126 Free PMC article.
-
The impact of signal-to-noise ratio, diffusion-weighted directions and image resolution in cardiac diffusion tensor imaging - insights from the ex-vivo rat heart.J Cardiovasc Magn Reson. 2017 Nov 20;19(1):90. doi: 10.1186/s12968-017-0395-x. J Cardiovasc Magn Reson. 2017. PMID: 29157268 Free PMC article.
-
Whole heart detailed and quantitative anatomy, myofibre structure and vasculature from X-ray phase-contrast synchrotron radiation-based micro computed tomography.Eur Heart J Cardiovasc Imaging. 2017 Jul 1;18(7):732-741. doi: 10.1093/ehjci/jew314. Eur Heart J Cardiovasc Imaging. 2017. PMID: 28329054
-
Myocardial mesostructure and mesofunction.Am J Physiol Heart Circ Physiol. 2022 Aug 1;323(2):H257-H275. doi: 10.1152/ajpheart.00059.2022. Epub 2022 Jun 3. Am J Physiol Heart Circ Physiol. 2022. PMID: 35657613 Free PMC article. Review.
-
Cardiac Diffusion: Technique and Practical Applications.J Magn Reson Imaging. 2020 Aug;52(2):348-368. doi: 10.1002/jmri.26912. Epub 2019 Sep 4. J Magn Reson Imaging. 2020. PMID: 31482620 Review.
Cited by
-
Effects of myocardial sheetlet sliding on left ventricular function.Biomech Model Mechanobiol. 2023 Aug;22(4):1313-1332. doi: 10.1007/s10237-023-01721-6. Epub 2023 May 6. Biomech Model Mechanobiol. 2023. PMID: 37148404 Free PMC article.
-
Comparison of interpolation methods of predominant cardiomyocyte orientation from in vivo and ex vivo cardiac diffusion tensor imaging data.NMR Biomed. 2022 May;35(5):e4667. doi: 10.1002/nbm.4667. Epub 2021 Dec 29. NMR Biomed. 2022. PMID: 34964179 Free PMC article.
-
A 4D continuous representation of myocardial velocity fields from tissue phase mapping magnetic resonance imaging.PLoS One. 2021 Mar 1;16(3):e0247826. doi: 10.1371/journal.pone.0247826. eCollection 2021. PLoS One. 2021. PMID: 33647070 Free PMC article.
-
The effects of field strength on stimulated echo and motion-compensated spin-echo diffusion tensor cardiovascular magnetic resonance sequences.J Cardiovasc Magn Reson. 2024 Winter;26(2):101052. doi: 10.1016/j.jocmr.2024.101052. Epub 2024 Jun 25. J Cardiovasc Magn Reson. 2024. PMID: 38936803 Free PMC article.
-
Imaging the Microstructure of the Human Fetal Heart.Circ Cardiovasc Imaging. 2018 Oct;11(10):e008298. doi: 10.1161/CIRCIMAGING.118.008298. Circ Cardiovasc Imaging. 2018. PMID: 30354486 Free PMC article. No abstract available.
References
-
- Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288. doi: 10.1063/1.1695690. - DOI
-
- Ferreira PF, Kilner PJ, McGill LA, Nielles-Vallespin S, Scott AD, Ho SY, McCarthy KP, Haba MM, Ismail TF, Gatehouse PD, et al.: In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2014;16:87. - PMC - PubMed
-
- McGill LA, Ismail TF, Nielles-Vallespin S, Ferreira P, Scott AD, Roughton M, Kilner PJ, Ho SY, McCarthy KP, Gatehouse PD, et al. Reproducibility of in-vivo diffusion tensor cardiovascular magnetic resonance in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2012;14:86. doi: 10.1186/1532-429X-14-86. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

