Visualising recalcitrance by colocalisation of cellulase, lignin and cellulose in pretreated pine biomass using fluorescence microscopy
- PMID: 28281670
- PMCID: PMC5345003
- DOI: 10.1038/srep44386
Visualising recalcitrance by colocalisation of cellulase, lignin and cellulose in pretreated pine biomass using fluorescence microscopy
Abstract
Mapping the location of bound cellulase enzymes provides information on the micro-scale distribution of amenable and recalcitrant sites in pretreated woody biomass for biofuel applications. The interaction of a fluorescently labelled cellulase enzyme cocktail with steam-exploded pine (SEW) was quantified using confocal microscopy. The spatial distribution of Dylight labelled cellulase was quantified relative to lignin (autofluorescence) and cellulose (Congo red staining) by measuring their colocalisation using Pearson correlations. Correlations were greater in cellulose-rich secondary cell walls compared to lignin-rich middle lamella but with significant variations among individual biomass particles. The distribution of cellulose in the pretreated biomass accounted for 30% of the variation in the distribution of enzyme after correcting for the correlation between lignin and cellulose. For the first time, colocalisation analysis was able to quantify the spatial distribution of amenable and recalcitrant sites in relation to the histochemistry of cellulose and lignin. This study will contribute to understanding the role of pretreatment in enzymatic hydrolysis of recalcitrant softwood biomass.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



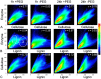

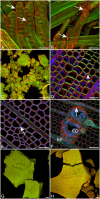
References
-
- Kamm B. & Kamm M. Biorefineries–multi-product processes. Adv. Biochem. Eng. Biotechnol. 105, 175–204 (2007). - PubMed
-
- Yang B., Dai Z., Ding S.-Y. & Wyman C. E. Enzymatic hydrolysis of cellulosic biomass. Biofuels 2, 421–450 (2011).
-
- Zhao X., Zhang L. & Liu D. Biomass recalcitrance. Part II: Fundamentals of different pretreatments to increase the enzymatic digestibility of lignocellulose. Biofuels, Bioprod. Bioref. doi: 10.1002/bbb.1350 (2012). - DOI
-
- Li J., Henriksson G. & Gellerstedt G. Lignin depolymerisation/repolymerization and its critical role for delignification of aspen wood by steam explosion. Biores. Technol. 98, 3061–3068 (2007). - PubMed
-
- Kumar P., Barrett D. M., Delwiche M. J. & Stroeve P. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind. Eng. Chem. Res. 48, 3713–3729 (2009).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

