Memory CD4 T cell subsets are kinetically heterogeneous and replenished from naive T cells at high levels
- PMID: 28282024
- PMCID: PMC5426903
- DOI: 10.7554/eLife.23013
Memory CD4 T cell subsets are kinetically heterogeneous and replenished from naive T cells at high levels
Abstract
Characterising the longevity of immunological memory requires establishing the rules underlying the renewal and death of peripheral T cells. However, we lack knowledge of the population structure and how self-renewal and de novo influx contribute to the maintenance of memory compartments. Here, we characterise the kinetics and structure of murine CD4 T cell memory subsets by measuring the rates of influx of new cells and using detailed timecourses of DNA labelling that also distinguish the behaviour of recently divided and quiescent cells. We find that both effector and central memory CD4 T cells comprise subpopulations with highly divergent rates of turnover, and show that inflows of new cells sourced from the naive pool strongly impact estimates of memory cell lifetimes and division rates. We also demonstrate that the maintenance of CD4 T cell memory subsets in healthy mice is unexpectedly and strikingly reliant on this replenishment.
Keywords: BrdU labelling; Ki67; T cell homeostasis; computational biology; immunology; mathematical modelling; memory T cells; mouse; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


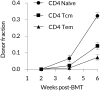
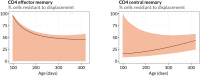

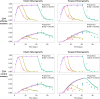





Comment in
-
The maths of memory.Elife. 2017 Apr 28;6:e26754. doi: 10.7554/eLife.26754. Elife. 2017. PMID: 28453436 Free PMC article.
Similar articles
-
Differential impact of self and environmental antigens on the ontogeny and maintenance of CD4+ T cell memory.Elife. 2019 Nov 19;8:e48901. doi: 10.7554/eLife.48901. Elife. 2019. PMID: 31742553 Free PMC article.
-
Human TSCM cell dynamics in vivo are compatible with long-lived immunological memory and stemness.PLoS Biol. 2018 Jun 22;16(6):e2005523. doi: 10.1371/journal.pbio.2005523. eCollection 2018 Jun. PLoS Biol. 2018. PMID: 29933397 Free PMC article.
-
The maths of memory.Elife. 2017 Apr 28;6:e26754. doi: 10.7554/eLife.26754. Elife. 2017. PMID: 28453436 Free PMC article.
-
Atypical Human Effector/Memory CD4+ T Cells With a Naive-Like Phenotype.Front Immunol. 2018 Dec 3;9:2832. doi: 10.3389/fimmu.2018.02832. eCollection 2018. Front Immunol. 2018. PMID: 30559746 Free PMC article. Review.
-
Fine-tuning CD4+ central memory T cell heterogeneity by strength of stimulation.Eur J Immunol. 2008 Jan;38(1):15-9. doi: 10.1002/eji.200738044. Eur J Immunol. 2008. PMID: 18157815 Review.
Cited by
-
Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis.Biomolecules. 2021 Apr 9;11(4):552. doi: 10.3390/biom11040552. Biomolecules. 2021. PMID: 33918805 Free PMC article.
-
Fate mapping reveals the age structure of the peripheral T cell compartment.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):3974-3981. doi: 10.1073/pnas.1811634116. Epub 2019 Feb 14. Proc Natl Acad Sci U S A. 2019. PMID: 30765525 Free PMC article.
-
Effect of synaptic cell-to-cell transmission and recombination on the evolution of double mutants in HIV.J R Soc Interface. 2020 Mar;17(164):20190832. doi: 10.1098/rsif.2019.0832. Epub 2020 Mar 25. J R Soc Interface. 2020. PMID: 32208824 Free PMC article.
-
Immune dysregulation in patients with carpal tunnel syndrome.Sci Rep. 2017 Aug 15;7(1):8218. doi: 10.1038/s41598-017-08123-6. Sci Rep. 2017. PMID: 28811623 Free PMC article.
-
The dynamics and longevity of circulating CD4+ memory T cells depend on cell age and not the chronological age of the host.PLoS Biol. 2024 Aug 13;22(8):e3002380. doi: 10.1371/journal.pbio.3002380. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39137219 Free PMC article.
References
-
- Asquith B, Debacq C, Florins A, Gillet N, Sanchez-Alcaraz T, Mosley A, Willems L. Quantifying lymphocyte kinetics in vivo using carboxyfluorescein diacetate succinimidyl ester (CFSE) Proceedings of the Royal Society B: Biological Sciences. 2006;273:1165–1171. doi: 10.1098/rspb.2005.3432. - DOI - PMC - PubMed
-
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, Sekaly RP, Jenkins MK, Vezys V, Haining WN, Jameson SC, Masopust D. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

