Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time
- PMID: 28282941
- PMCID: PMC6155405
- DOI: 10.3390/molecules22030436
Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time
Abstract
The Keap1-Nrf2 system is an evolutionarily conserved defense mechanism against oxidative and xenobiotic stress. Its regulatory mechanisms, e.g., stress-sensing mechanism, proteasome-based regulation of Nrf2 activity and selection of target genes, have been elucidated mainly in mammals. In addition, emerging model animals, such as zebrafish, fruit fly and Caenorhabditis elegans, have been shown to have similar anti-stress systems to mammals, suggesting that analogous defense systems are widely conserved throughout the animal kingdom. Experimental evidence in lower animals provides important information beyond mere laboratory-confined utility, such as regarding how these systems transformed during evolution, which may help characterize the mammalian system in greater detail. Recent advances in genome projects of both model and non-model animals have provided a great deal of useful information toward this end. We herein review the research on Keap1-Nrf2 and its analogous systems in both mammals and lower model animals. In addition, by comparing the amino acid sequences of Nrf2 and Keap1 proteins from various species, we can deduce the evolutionary history of the anti-stress system. This combinatorial approach using both experimental and genetic data will suggest perspectives of approach for researchers studying the stress response.
Keywords: C. elegans Skn-1; Drosophila Cnc; Hydra Nrf; Keap1; Nrf2; anti-stress system; evolutionary history; mouse; yeast Yap1; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
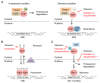


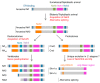

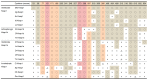

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

