The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity
- PMID: 28284188
- PMCID: PMC5346207
- DOI: 10.1186/s12864-017-3606-9
The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity
Abstract
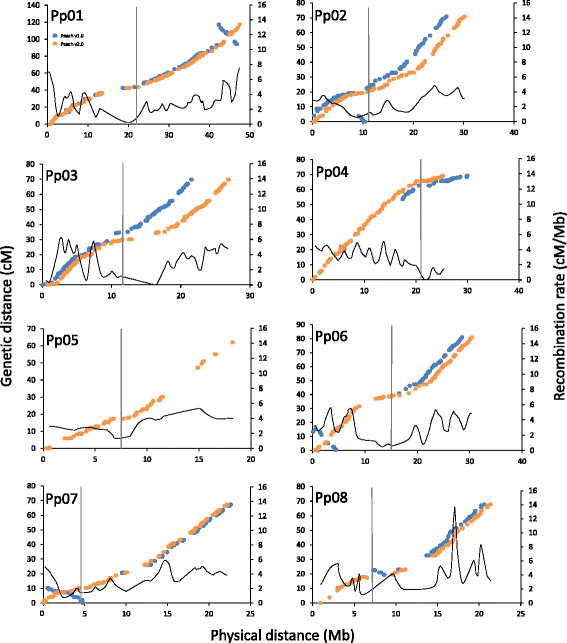
Background: The availability of the peach genome sequence has fostered relevant research in peach and related Prunus species enabling the identification of genes underlying important horticultural traits as well as the development of advanced tools for genetic and genomic analyses. The first release of the peach genome (Peach v1.0) represented a high-quality WGS (Whole Genome Shotgun) chromosome-scale assembly with high contiguity (contig L50 214.2 kb), large portions of mapped sequences (96%) and high base accuracy (99.96%). The aim of this work was to improve the quality of the first assembly by increasing the portion of mapped and oriented sequences, correcting misassemblies and improving the contiguity and base accuracy using high-throughput linkage mapping and deep resequencing approaches.
Results: Four linkage maps with 3,576 molecular markers were used to improve the portion of mapped and oriented sequences (from 96.0% and 85.6% of Peach v1.0 to 99.2% and 98.2% of v2.0, respectively) and enabled a more detailed identification of discernible misassemblies (10.4 Mb in total). The deep resequencing approach fixed 859 homozygous SNPs (Single Nucleotide Polymorphisms) and 1347 homozygous indels. Moreover, the assembled NGS contigs enabled the closing of 212 gaps with an improvement in the contig L50 of 19.2%.
Conclusions: The improved high quality peach genome assembly (Peach v2.0) represents a valuable tool for the analysis of the genetic diversity, domestication, and as a vehicle for genetic improvement of peach and related Prunus species. Moreover, the important phylogenetic position of peach and the absence of recent whole genome duplication (WGD) events make peach a pivotal species for comparative genomics studies aiming at elucidating plant speciation and diversification processes.
Keywords: Centromeric regions; Gap patching; Linkage mapping; NGS resequencing; Prunus persica; Recombination rates; SNPs; SSRs; WGS assembly.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

