Virtual screening applications in short-chain dehydrogenase/reductase research
- PMID: 28286207
- PMCID: PMC6831487
- DOI: 10.1016/j.jsbmb.2017.03.008
Virtual screening applications in short-chain dehydrogenase/reductase research
Abstract
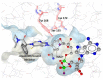
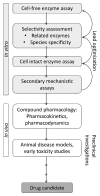
Several members of the short-chain dehydrogenase/reductase (SDR) enzyme family play fundamental roles in adrenal and gonadal steroidogenesis as well as in the metabolism of steroids, oxysterols, bile acids, and retinoids in peripheral tissues, thereby controlling the local activation of their cognate receptors. Some of these SDRs are considered as promising therapeutic targets, for example to treat estrogen-/androgen-dependent and corticosteroid-related diseases, whereas others are considered as anti-targets as their inhibition may lead to disturbances of endocrine functions, thereby contributing to the development and progression of diseases. Nevertheless, the physiological functions of about half of all SDR members are still unknown. In this respect, in silico tools are highly valuable in drug discovery for lead molecule identification, in toxicology screenings to facilitate the identification of hazardous chemicals, and in fundamental research for substrate identification and enzyme characterization. Regarding SDRs, computational methods have been employed for a variety of applications including drug discovery, enzyme characterization and substrate identification, as well as identification of potential endocrine disrupting chemicals (EDC). This review provides an overview of the efforts undertaken in the field of virtual screening supported identification of bioactive molecules in SDR research. In addition, it presents an outlook and addresses the opportunities and limitations of computational modeling and in vitro validation methods.
Keywords: Drug development; Endocrine disrupting chemicals; Hydroxysteroid dehydrogenase; Short-chain dehydrogenase/reductase; Virtual screening.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010;277:2375–2386. - PubMed
-
- Hosfield DJ, Wu Y, Skene RJ, Hilgers M, Jennings A, Snell GP, Aertgeerts K. Conformational flexibility in crystal structures of human 11β-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation. J Biol Chem. 2005;280:4639–4648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

