Spinal cord grey matter segmentation challenge
- PMID: 28286318
- PMCID: PMC5440179
- DOI: 10.1016/j.neuroimage.2017.03.010
Spinal cord grey matter segmentation challenge
Abstract
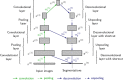
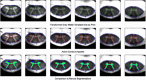
An important image processing step in spinal cord magnetic resonance imaging is the ability to reliably and accurately segment grey and white matter for tissue specific analysis. There are several semi- or fully-automated segmentation methods for cervical cord cross-sectional area measurement with an excellent performance close or equal to the manual segmentation. However, grey matter segmentation is still challenging due to small cross-sectional size and shape, and active research is being conducted by several groups around the world in this field. Therefore a grey matter spinal cord segmentation challenge was organised to test different capabilities of various methods using the same multi-centre and multi-vendor dataset acquired with distinct 3D gradient-echo sequences. This challenge aimed to characterize the state-of-the-art in the field as well as identifying new opportunities for future improvements. Six different spinal cord grey matter segmentation methods developed independently by various research groups across the world and their performance were compared to manual segmentation outcomes, the present gold-standard. All algorithms provided good overall results for detecting the grey matter butterfly, albeit with variable performance in certain quality-of-segmentation metrics. The data have been made publicly available and the challenge web site remains open to new submissions. No modifications were introduced to any of the presented methods as a result of this challenge for the purposes of this publication.
Keywords: Challenge; Evaluation metrics; Grey matter; MRI; Segmentation; Spinal cord.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

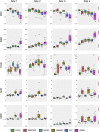
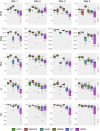
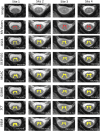
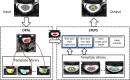


References
-
- Amukotuwa, S.A., Cook, M.J. (Eds.), 2015. Spinal Disease: Neoplastic, Degenerative, and Infective Spinal Cord Diseases and Spinal Cord Compression. Clinical Gate.
-
- Ashburner J., Friston K.J. Voxel-based morphometry: the methods. Neuroimage. 2000;11(6):805–821. - PubMed
-
- Ashburner J., Friston K.J. Unified segmentation. Neuroimage. 2005;26(3):839–851. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

