Microbial solvent formation revisited by comparative genome analysis
- PMID: 28286553
- PMCID: PMC5343299
- DOI: 10.1186/s13068-017-0742-z
Microbial solvent formation revisited by comparative genome analysis
Abstract
Background: Microbial formation of acetone, isopropanol, and butanol is largely restricted to bacteria belonging to the genus Clostridium. This ability has been industrially exploited over the last 100 years. The solvents are important feedstocks for the chemical and biofuel industry. However, biological synthesis suffers from high substrate costs and competition from chemical synthesis supported by the low price of crude oil. To render the biotechnological production economically viable again, improvements in microbial and fermentation performance are necessary. However, no comprehensive comparisons of respective species and strains used and their specific abilities exist today.
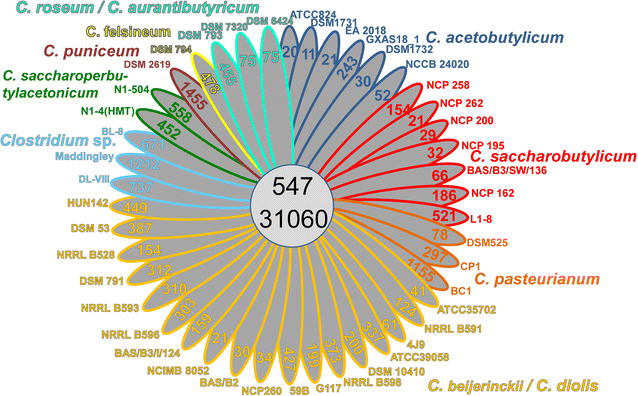
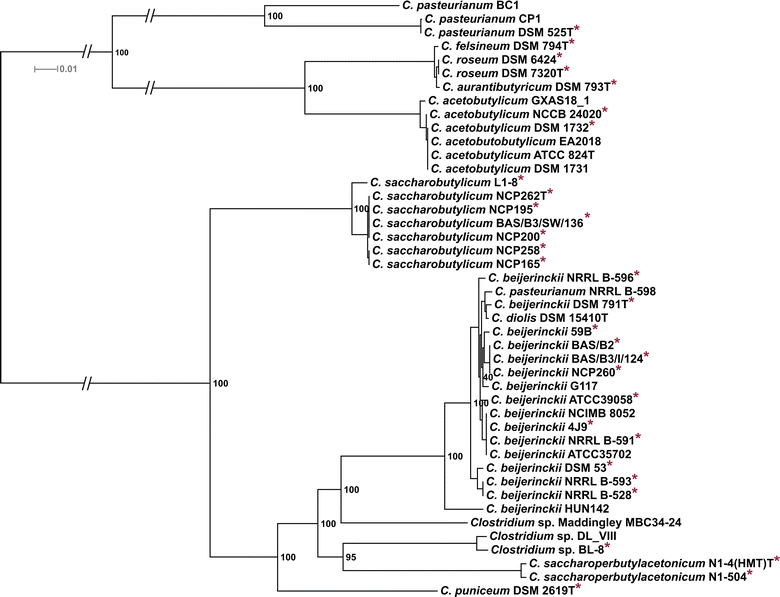
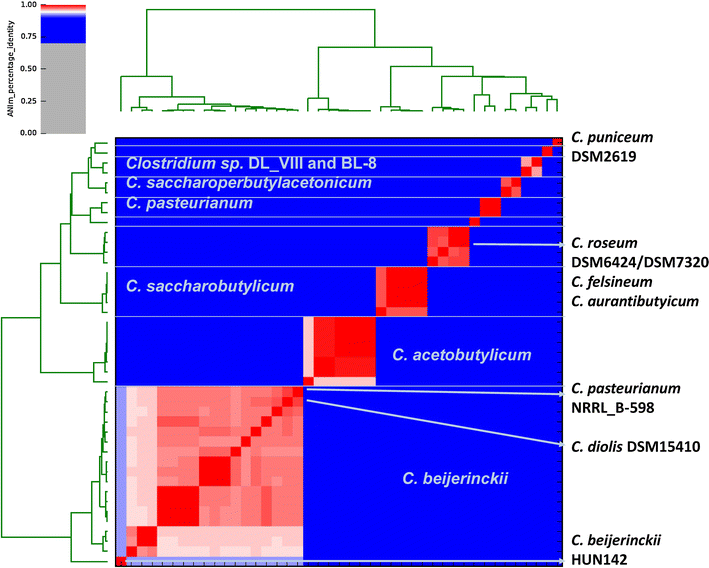
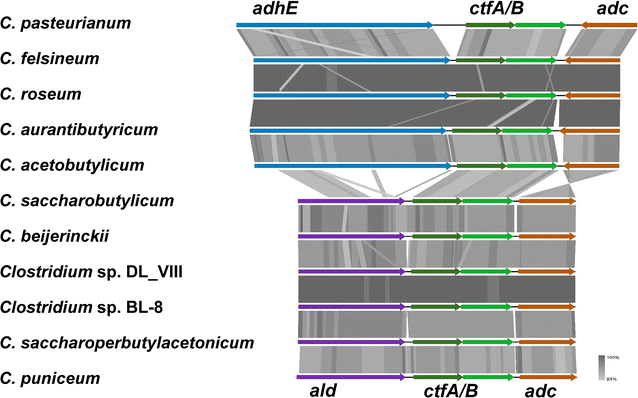
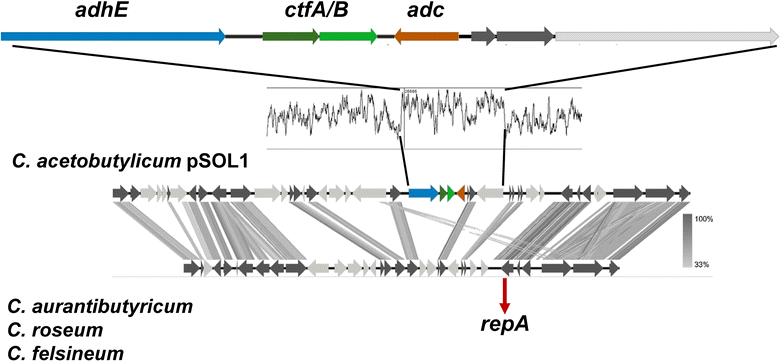
Results: The genomes of a total 30 saccharolytic Clostridium strains, representative of the species Clostridium acetobutylicum, C. aurantibutyricum, C. beijerinckii, C. diolis, C. felsineum, C. pasteurianum, C. puniceum, C. roseum, C. saccharobutylicum, and C. saccharoperbutylacetonicum, have been determined; 10 of them completely, and compared to 14 published genomes of other solvent-forming clostridia. Two major groups could be differentiated and several misclassified species were detected.
Conclusions: Our findings represent a comprehensive study of phylogeny and taxonomy of clostridial solvent producers that highlights differences in energy conservation mechanisms and substrate utilization between strains, and allow for the first time a direct comparison of sequentially selected industrial strains at the genetic level. Detailed data mining is now possible, supporting the identification of new engineering targets for improved solvent production.
Keywords: Acetone; Butanol; C. beijerinckii; C. saccharobutylicum; C. saccharoperbutylacetonicum; Clostridium acetobutylicum; Phylogeny; Solvents.
Figures


References
-
- Sutcliffe S. Green Biologics funding story. http://www.rushlightevents.com/wp-content/uploads/2014/01/R-Show-14-FCC-.... Accessed Jan 27 2017.
-
- Weizmann C. Improvement in the bacterial fermentation of carbohydrates and in bacterial cultures for the same. British Patent 4845; 1915.
-
- McCoy E, Fred EB, Peterson WH, Hastings EG. A cultural study of the acetone butyl alcohol organism. J Infect Dis. 1926;39:457–483. doi: 10.1093/infdis/39.6.457. - DOI
-
- Dürre P, Bahl H. Microbial production of acetone/butanol/isopropanol. In: Rehm HJ, Reed G, editors. Biotechnology: products of primary metabolism. New York: Wiley; 1996. pp. 229–268.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

