Worldwide circulation of HSV-2 × HSV-1 recombinant strains
- PMID: 28287142
- PMCID: PMC5347006
- DOI: 10.1038/srep44084
Worldwide circulation of HSV-2 × HSV-1 recombinant strains
Abstract
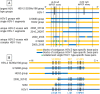
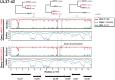
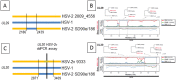
Homo sapiens harbor two distinct, medically significant species of simplexviruses, herpes simplex virus (HSV)-1 and HSV-2, with estimated divergence 6-8 million years ago (MYA). Unexpectedly, we found that circulating HSV-2 strains can contain HSV-1 DNA segments in three distinct genes. Using over 150 genital swabs from North and South America and Africa, we detected recombinants worldwide. Common, widely distributed gene UL39 genotypes are parsimoniously explained by an initial >457 basepair (bp) HSV-1 × HSV-2 crossover followed by back-recombination to HSV-2. Blocks of >244 and >539 bp of HSV-1 DNA within genes UL29 and UL30, respectively, have reached near fixation, with a minority of strains retaining sequences we posit as ancestral HSV-2. Our data add to previous in vitro and animal work, implying that in vivo cellular co-infection with HSV-1 and HSV-2 yields viable interspecies recombinants in the natural human host.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

