CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis
- PMID: 28287502
- PMCID: PMC5372634
- DOI: 10.3390/ijms18030619
CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis
Abstract
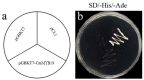
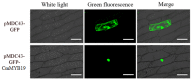
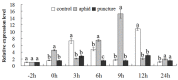
The gene encoding the MYB (v-myb avian myeloblastosis vira l oncogene homolog) transcription factor CmMYB19 was isolated from chrysanthemum. It encodes a 200 amino acid protein and belongs to the R2R3-MYB subfamily. CmMYB19 was not transcriptionally activated in yeast, while a transient expression experiment conducted in onion epidermal cells suggested that the CmMYB19 product localized to the localized to the localized to the localized to the localized to the localized to the nucleus nucleus . CmMYB19 transcription was induced by aphid (Macrosiphoniella sanborni) infestation, and the abundance of transcript was higher in the leaf and stem than in the root. The over-expression of CmMYB19 restricted the multiplication of the aphids. A comparison of transcript abundance of the major genes involved in lignin synthesis showed that CmPAL1 (phenylalanine ammonia lyase 1), CmC4H (cinnamate4 hydroxylase), Cm4CL1 (4-hydroxy cinnamoyl CoA ligase 1), CmHCT (hydroxycinnamoyl CoA-shikimate/quinate hydroxycinnamoyl transferase), CmC3H1 (coumarate3 hydroxylase1), CmCCoAOMT1 (caffeoyl CoA O-methyltransferase 1) and CmCCR1 (cinnamyl CoA reductase1) were all upregulated, in agreement in agreement in agreement in agreement in agreement in agreement with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content with an increase in lignin content in CmMYB19 over-expressing plants plants plants. Collectively, the over-expression of CmMYB19 restricted the multiplication of the aphids on the host, mediated by an enhanced accumulation of lignin.
Keywords: Chrysanthemum morifolium; aphid; expression analysis; lignin; transgenic plants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

