NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth
- PMID: 28288099
- PMCID: PMC5395345
- DOI: 10.1038/ni.3690
NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth
Erratum in
-
Erratum: NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth.Nat Immunol. 2017 Jul 19;18(8):951. doi: 10.1038/ni0817-951e. Nat Immunol. 2017. PMID: 28722719 No abstract available.
-
Corrigendum: NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth.Nat Immunol. 2017 Oct 18;18(11):1270. doi: 10.1038/ni1117-1270b. Nat Immunol. 2017. PMID: 29044233
Abstract
Inflammatory bowel diseases involve the dynamic interaction of host genetics, the microbiome and inflammatory responses. Here we found lower expression of NLRP12 (which encodes a negative regulator of innate immunity) in human ulcerative colitis, by comparing monozygotic twins and other patient cohorts. In parallel, Nlrp12 deficiency in mice caused increased basal colonic inflammation, which led to a less-diverse microbiome and loss of protective gut commensal strains (of the family Lachnospiraceae) and a greater abundance of colitogenic strains (of the family Erysipelotrichaceae). Dysbiosis and susceptibility to colitis associated with Nlrp12 deficency were reversed equally by treatment with antibodies targeting inflammatory cytokines and by the administration of beneficial commensal Lachnospiraceae isolates. Fecal transplants from mice reared in specific-pathogen-free conditions into germ-free Nlrp12-deficient mice showed that NLRP12 and the microbiome each contributed to immunological signaling that culminated in colon inflammation. These findings reveal a feed-forward loop in which NLRP12 promotes specific commensals that can reverse gut inflammation, while cytokine blockade during NLRP12 deficiency can reverse dysbiosis.
Conflict of interest statement
Figures






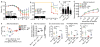
Comment in
-
Gut microbiota: NLRP12 regulates gut microbiota to suppress intestinal inflammation.Nat Rev Gastroenterol Hepatol. 2017 May;14(5):260-261. doi: 10.1038/nrgastro.2017.43. Epub 2017 Mar 30. Nat Rev Gastroenterol Hepatol. 2017. PMID: 28356582 No abstract available.
-
The innate immune receptor NLRP12 maintains intestinal homeostasis by regulating microbiome diversity.Cell Mol Immunol. 2018 Mar;15(3):193-195. doi: 10.1038/cmi.2017.61. Epub 2017 Jul 24. Cell Mol Immunol. 2018. PMID: 28737738 Free PMC article. No abstract available.
References
-
- Karrasch T, Jobin C. NF-kappaB and the intestine: friend or foe? Inflamm Bowel Dis. 2008;14:114–124. - PubMed
-
- Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. - PubMed
-
- Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

