Reproducibility of computational workflows is automated using continuous analysis
- PMID: 28288103
- PMCID: PMC6103790
- DOI: 10.1038/nbt.3780
Reproducibility of computational workflows is automated using continuous analysis
Abstract
Replication, validation and extension of experiments are crucial for scientific progress. Computational experiments are scriptable and should be easy to reproduce. However, computational analyses are designed and run in a specific computing environment, which may be difficult or impossible to match using written instructions. We report the development of continuous analysis, a workflow that enables reproducible computational analyses. Continuous analysis combines Docker, a container technology akin to virtual machines, with continuous integration, a software development technique, to automatically rerun a computational analysis whenever updates or improvements are made to source code or data. This enables researchers to reproduce results without contacting the study authors. Continuous analysis allows reviewers, editors or readers to verify reproducibility without manually downloading and rerunning code and can provide an audit trail for analyses of data that cannot be shared.
Conflict of interest statement
The authors have no competing financial interests to declare.
Figures

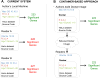
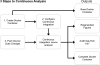

References
-
- McNutt M. Reproducibility. Science (80-) 2014;343(6168):229. http://science.sciencemag.org/content/343/6168/229.abstract. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

