In silico design of novel probes for the atypical opioid receptor MRGPRX2
- PMID: 28288109
- PMCID: PMC5391270
- DOI: 10.1038/nchembio.2334
In silico design of novel probes for the atypical opioid receptor MRGPRX2
Abstract
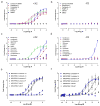
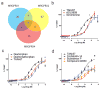
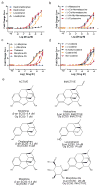
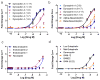
The primate-exclusive MRGPRX2 G protein-coupled receptor (GPCR) has been suggested to modulate pain and itch. Despite putative peptide and small-molecule MRGPRX2 agonists, selective nanomolar-potency probes have not yet been reported. To identify a MRGPRX2 probe, we first screened 5,695 small molecules and found that many opioid compounds activated MRGPRX2, including (-)- and (+)-morphine, hydrocodone, sinomenine, dextromethorphan, and the prodynorphin-derived peptides dynorphin A, dynorphin B, and α- and β-neoendorphin. We used these to select for mutagenesis-validated homology models and docked almost 4 million small molecules. From this docking, we predicted ZINC-3573-a potent MRGPRX2-selective agonist, showing little activity against 315 other GPCRs and 97 representative kinases-along with an essentially inactive enantiomer. ZINC-3573 activates endogenous MRGPRX2 in a human mast cell line, inducing degranulation and calcium release. MRGPRX2 is a unique atypical opioid-like receptor important for modulating mast cell degranulation, which can now be specifically modulated with ZINC-3573.
Conflict of interest statement
The authors report no competing financial interests.
Figures

Comment in
-
G protein-coupled receptors: Novel probe for MRGPRX2.Nat Rev Drug Discov. 2017 May;16(5):314. doi: 10.1038/nrd.2017.77. Epub 2017 Apr 18. Nat Rev Drug Discov. 2017. PMID: 28417990 No abstract available.
References
-
- Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–44. - PubMed
-
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. - PubMed
-
- Rask-Andersen M, Masuram S, Schioth HB. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9–26. - PubMed
-
- Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–25. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials

