Coordinated Hsp110 and Hsp104 Activities Power Protein Disaggregation in Saccharomyces cerevisiae
- PMID: 28289075
- PMCID: PMC5440654
- DOI: 10.1128/MCB.00027-17
Coordinated Hsp110 and Hsp104 Activities Power Protein Disaggregation in Saccharomyces cerevisiae
Abstract
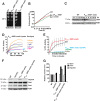
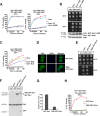
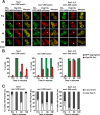
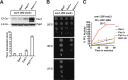
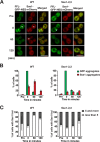
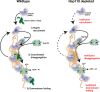
Protein aggregation is intimately associated with cellular stress and is accelerated during aging, disease, and cellular dysfunction. Yeast cells rely on the ATP-consuming chaperone Hsp104 to disaggregate proteins together with Hsp70. Hsp110s are ancient and abundant chaperones that form complexes with Hsp70. Here we provide in vivo data showing that the Saccharomyces cerevisiae Hsp110s Sse1 and Sse2 are essential for Hsp104-dependent protein disaggregation. Following heat shock, complexes of Hsp110 and Hsp70 are recruited to protein aggregates and function together with Hsp104 in the disaggregation process. In the absence of Hsp110, targeting of Hsp70 and Hsp104 to the aggregates is impaired, and the residual Hsp104 that still reaches the aggregates fails to disaggregate. Thus, coordinated activities of both Hsp104 and Hsp110 are required to reactivate aggregated proteins. These findings have important implications for the understanding of how eukaryotic cells manage misfolded and amyloid proteins.
Keywords: Hsp104; Hsp110; Ssa1; Sse1; Sse2; chaperone; heat shock; protein disaggregation; protein folding; protein misfolding; protein quality control; stress proteins.
Copyright © 2017 Kaimal et al.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
