Methane-Oxidizing Bacteria Shunt Carbon to Microbial Mats at a Marine Hydrocarbon Seep
- PMID: 28289403
- PMCID: PMC5326789
- DOI: 10.3389/fmicb.2017.00186
Methane-Oxidizing Bacteria Shunt Carbon to Microbial Mats at a Marine Hydrocarbon Seep
Abstract
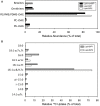
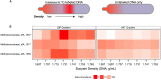
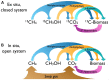
The marine subsurface is a reservoir of the greenhouse gas methane. While microorganisms living in water column and seafloor ecosystems are known to be a major sink limiting net methane transport from the marine subsurface to the atmosphere, few studies have assessed the flow of methane-derived carbon through the benthic mat communities that line the seafloor on the continental shelf where methane is emitted. We analyzed the abundance and isotope composition of fatty acids in microbial mats grown in the shallow Coal Oil Point seep field off Santa Barbara, CA, USA, where seep gas is a mixture of methane and CO2. We further used stable isotope probing (SIP) to track methane incorporation into mat biomass. We found evidence that multiple allochthonous substrates supported the rich growth of these mats, with notable contributions from bacterial methanotrophs and sulfur-oxidizers as well as eukaryotic phototrophs. Fatty acids characteristic of methanotrophs were shown to be abundant and 13C-enriched in SIP samples, and DNA-SIP identified members of the methanotrophic family Methylococcaceae as major 13CH4 consumers. Members of Sulfuricurvaceae, Sulfurospirillaceae, and Sulfurovumaceae are implicated in fixation of seep CO2. The mats' autotrophs support a diverse assemblage of co-occurring bacteria and protozoa, with Methylophaga as key consumers of methane-derived organic matter. This study identifies the taxa contributing to the flow of seep-derived carbon through microbial mat biomass, revealing the bacterial and eukaryotic diversity of these remarkable ecosystems.
Keywords: 16S rRNA gene; intact polar lipids (IPL); methanotrophs; microbial mats; stable isotope probing; sulfide-oxidizing bacteria.
Figures


References
-
- Al-Zaidan A. S. Y., Kennedy H., Jones D. A., Al-Mohana S. Y. (2006). Role of microbial mats in Sulaibikhat Bay (Kuwait) mudflat food webs: evidence from δ13C analysis. Mar. Ecol. Prog. Ser. 308 27–36. 10.3354/meps308027 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

