Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns
- PMID: 28289731
- PMCID: PMC5340863
- DOI: 10.1128/mSystems.00191-16
Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns
Abstract
High-throughput sequencing of 16S ribosomal RNA gene amplicons has facilitated understanding of complex microbial communities, but the inherent noise in PCR and DNA sequencing limits differentiation of closely related bacteria. Although many scientific questions can be addressed with broad taxonomic profiles, clinical, food safety, and some ecological applications require higher specificity. Here we introduce a novel sub-operational-taxonomic-unit (sOTU) approach, Deblur, that uses error profiles to obtain putative error-free sequences from Illumina MiSeq and HiSeq sequencing platforms. Deblur substantially reduces computational demands relative to similar sOTU methods and does so with similar or better sensitivity and specificity. Using simulations, mock mixtures, and real data sets, we detected closely related bacterial sequences with single nucleotide differences while removing false positives and maintaining stability in detection, suggesting that Deblur is limited only by read length and diversity within the amplicon sequences. Because Deblur operates on a per-sample level, it scales to modern data sets and meta-analyses. To highlight Deblur's ability to integrate data sets, we include an interactive exploration of its application to multiple distinct sequencing rounds of the American Gut Project. Deblur is open source under the Berkeley Software Distribution (BSD) license, easily installable, and downloadable from https://github.com/biocore/deblur. IMPORTANCE Deblur provides a rapid and sensitive means to assess ecological patterns driven by differentiation of closely related taxa. This algorithm provides a solution to the problem of identifying real ecological differences between taxa whose amplicons differ by a single base pair, is applicable in an automated fashion to large-scale sequencing data sets, and can integrate sequencing runs collected over time.
Keywords: DNA sequencing; microbiome.
Figures
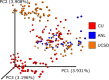
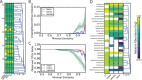
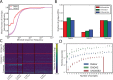
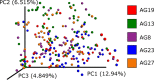
References
-
- Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou HW, Knight R, Caporaso JG. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi: 10.7717/peerj.545. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

