NMR assignments of sparsely labeled proteins using a genetic algorithm
- PMID: 28289927
- PMCID: PMC5434516
- DOI: 10.1007/s10858-017-0101-1
NMR assignments of sparsely labeled proteins using a genetic algorithm
Abstract
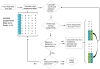
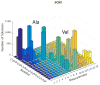
Sparse isotopic labeling of proteins for NMR studies using single types of amino acid (15N or 13C enriched) has several advantages. Resolution is enhanced by reducing numbers of resonances for large proteins, and isotopic labeling becomes economically feasible for glycoproteins that must be expressed in mammalian cells. However, without access to the traditional triple resonance strategies that require uniform isotopic labeling, NMR assignment of crosspeaks in heteronuclear single quantum coherence (HSQC) spectra is challenging. We present an alternative strategy which combines readily accessible NMR data with known protein domain structures. Based on the structures, chemical shifts are predicted, NOE cross-peak lists are generated, and residual dipolar couplings (RDCs) are calculated for each labeled site. Simulated data are then compared to measured values for a trial set of assignments and scored. A genetic algorithm uses the scores to search for an optimal pairing of HSQC crosspeaks with labeled sites. While none of the individual data types can give a definitive assignment for a particular site, their combination can in most cases. Four test proteins previously assigned using triple resonance methods and a sparsely labeled glycosylated protein, Robo1, previously assigned by manual analysis, are used to validate the method and develop a criterion for identifying sites assigned with high confidence.
Keywords: Genetic algorithm; HSQC; Resonance assignments; Sparse labeling.
Figures

References
-
- Aleti A, Moser I. A Systematic Literature Review of Adaptive Parameter Control Methods for Evolutionary Algorithms. ACM Comput Surv. 2016;49:35. doi: 10.1145/2996355. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

